DEEP EHR: CHRONIC DISEASE PREDICTION USING MEDICAL NOTES
4. Methods
4.1 Baselines
We use three different baselines to evaluate the contribution of text data, the deep learning architecture
and their combination. Firstly, we train an L1-regularized logistic regression model with all available
demographic features and lab values averaged within the history window. Secondly we train an
LSTM model with all available demographic features and lab values at each encounter, to provide a
more fair comparison with other deep learning models with additional text data. The last baseline is
an L1-regularized logistic regression with TF-IDF N-gram features in the text. This baseline shows
the value of deep learning in modeling complicated text. We use the 20k most frequent 1-, 2-, and
3-grams. We utilize the sklearn’s implementation of logistic regression for this task Pedregosa et al.
(2011).
4.2 Learning Continuous Embedding of Vocabulary
In our first analysis we use embeddings previously trained on the PubMed dataset Pyysalo and
Ananiadou (2013). We then train new embeddings directly on the NYU Langone Center medical
notes, as the style and abbreviations present in clinical notes are distinct from medical publications
available at PubMed. We adopt StarSpace Wu et al. (2017) as a general-purpose neural model for
efficient learning of entity embeddings. In particular, we label the notes from each encounter with
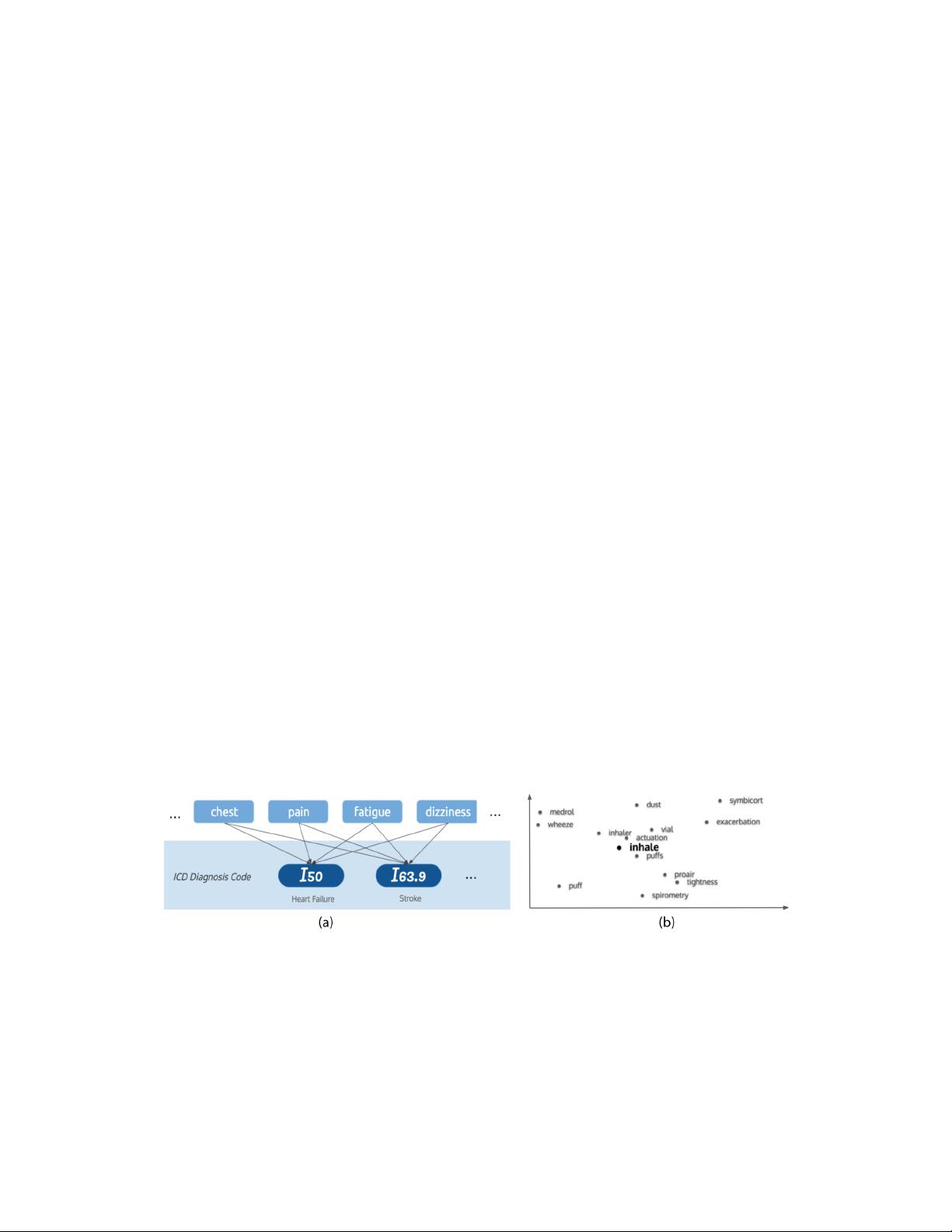
the ICD-10 diagnosis codes of the same encounter, as shown in Figure 2 (a). Under StarSpace’s
bag-of-word approach, the encounter is represented by aggregating the embedding of individual
words (we used the default aggregation method where the encounter is the sum of embeddings of
all words divided by the squared root of number of words). Both the word embeddings and the
diagnosis code embeddings are trained so that the cosine similarity between the encounter and its
diagnoses is ranked higher than that between the encounter and a set of different diagnoses. Thus
words related to the same symptom are placed close to each other in the embedding space. For
example, figure 2 (b) shows neighbours of the word "inhale" by t-SNE projection of the embeddings
to the 2-dimensional space. We find that the bag-of-word style embedding creates representations
better for disease prediction than using a standard skip-gram objective.
Figure 2: Illustration of StarSpace: (a) embeddings are trained with labeled bag-of-words approach.
(b) StarSpace word embedding example under t-SNE projection.
Utilizing all available notes except for those on patients in the validation or the test set, we
obtain a much larger embedding training set than that of the prediction task. We find that the
StarSpace embeddings trained on clinical notes outperform the pre-trained PubMed embeddings in
the downstream prediction task, as shown in Table 2. We test a set of 300-dimension embeddings and
5









