522 | Mol. BioSyst., 2016, 12, 520--531 This journal is
©
The Royal Society of Chemistry 2016
heterogeneous network. Thirdly, implementation of label pro-
pagation on the drug (or target) similarity sub-network to
obtain the drug (or target) label network. Fourthly, implemen-
tation of label propagation on the target (or drug) similarity
sub-network, whose initial label information is derived from
the drug (or target) label network and the drug–target bipartite
network. Finally, the most probable targets (or drugs) are
selected according to the stable label scores of the walk.
LPMIHN is mainly different from NRWRH in three aspects.
One is that the drug/target similarity network integrates the
topological information of the known drug–target interaction
network. Another is that label propagation (or random walk) is
implemented on the drug and target similarity networks,
respectively. Thirdly, the initial label information of the target/
drug network comes from the drug/target label network and the
known drug–target bipartite network.
Through extensive simulations on four benchmark datasets
and two quantitative kinase bioactivity datasets, LPMIHN
shows better performance than the existing state-of-the-art
methods, such as BLM-NII, NetCBP and NRWRH. Furthermore,
some new predicted drug–target interactions ranked in top
were reported by publicly accessible datasets. It is anticipated
that our LPMIHN algorithm can help us to find new or potential
drug–target interactions, and provide useful information for
drug design.
2 Materials
To facilitate benchmarking comparison with other state-of-art
methods, we used the four drug–target interaction datasets
from humans, namely enzymes (Es), ion channels (ICs),
G-protein coupled receptors (GPCRs) and nuclear receptors
(NRs), which were originally provided by Yamanishi et al.,
40
and widely used as the benchmark binary interaction datasets
of compounds targeting pharmaceutically useful target pro-
teins.
29,31,34,35,42–44,47,48
These datasets are available at http://
web.kuicr.kyoto-u.ac.jp/supp/yoshi/drugtarget/. The Es dataset
includes 445 drugs, 664 targets and 2926 known drug–target
interactions. The ICs dataset includes 210 drugs, 204 targets
and 1476 known drug–target interactions. The GPCRs dataset
includes 223 drugs, 95 targets and 635 known drug–target
interactions. The NRs dataset includes 54 drugs, 26 targets
and 90 known drug–target interactions.
As binary interaction datasets ignore many important
characteristics of the drug–target interaction, such as dose-
dependence and quantitative affinity, we use the same cutoff
thresholds of K
d
r 30.00 nM and K
i
o 28.18 nM as ref. 18 to
binarize two large-scale quantitative kinase bioactivity datasets,
i.e., kinase disassociation constant (K
d
) dataset and kinase
inhibition constant (K
i
) dataset,
49,50
forming two binary inter-
action datasets which include 68 drugs, 442 targets and 1527
drug–target interactions for the K
d
dataset, and 1421 drugs, 156
targets and 3200 drug–target interactions for the K
i
dataset.
These two datasets are applied to evaluate the performance of
our LPMIHN algorithm. The smaller the K
d
/K
i
bioactivity, the
higher the interaction affinity between the chemical compound
and the protein kinase.
Table 1 lists some statistics of each dataset including the
total number of drugs (N
d
), the total number of targets (N
t
), the
total number of interaction edges (E
dt
), the total number of
drugs that have only one targeting protein (k
d
(1)), the total
number of targets that have only one associated drug (k
t
(1)), the
average number of targets for each drug (avg. N
d
), the average
number of drugs for each target (avg. N
t
), and the sparsity
which is defined as the total number of connected edges in the
real network divided by the total number of linked edges in the
complete graph.
3 Methods
Our LPMIHN method can be divided into two parts: construct-
ing the heterogeneous network and separately implementing
label propagation on the drug/target similarity networks.
3.1 Heterogeneous network
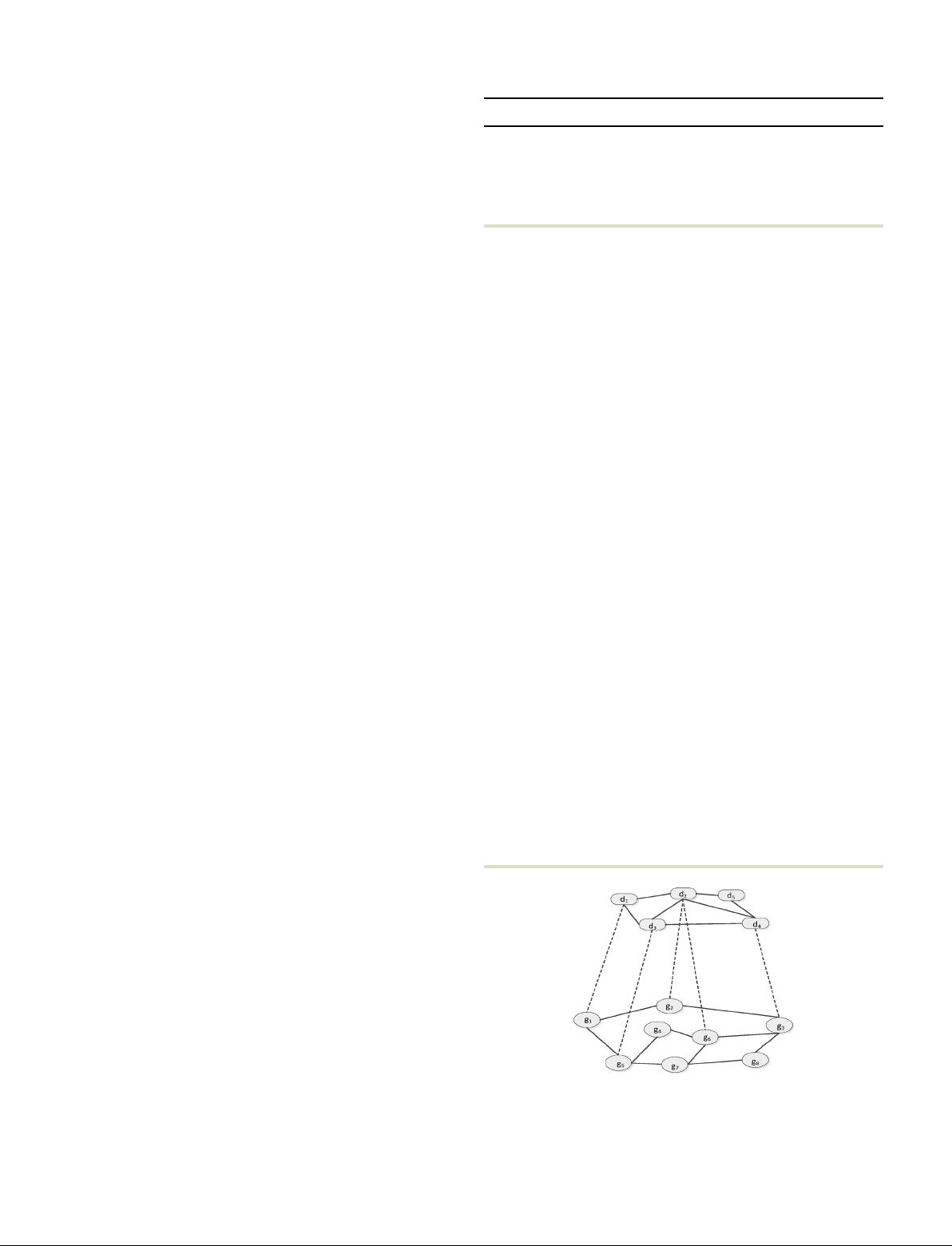
The heterogeneous network of drug–target interactions is
composed of three typical networks: the drug similarity net-
work, target similarity network and the known drug–target
interaction bipartite graph network (see Fig. 1).
The matrix S
d
corresponding to the drug similarity network
is composed of the chemical structure similarity matrix S
c
d
and
the drug–target interaction profile-based drug similarity matrix
S
IP
d
. The matrix S
g
corresponding to the target protein similarity
network is composed of the protein sequence similarity matrix
S
s
g
and the drug–target interaction profile-based target similarity
matrix S
IP
g
. The drug–target interaction adjacent matrix A
Table 1 Statistical characteristics of six drug–target interaction datasets
Dataset N
d
N
t
E
dt
k
d
(1) k
t
(1) avg. N
d
avg. N
t
Sparsity
Es 445 664 2926 177 288 6.58 4.41 0.0099
ICs 210 204 1476 81 23 7.03 7.24 0.0344
GPCRs 223 95 635 106 34 2.85 6.68 0.0299
NRs 54 26 90 39 8 1.67 3.46 0.0641
K
d
68 442 1527 4 97 22.46 3.45 0.0508
K
i
1421 156 3200 204 11 2.25 20.51 0.0144
Fig. 1 Drug–target interaction heterogeneous network model. The upper
sub-network is the drug similarity network, the underlying sub-network is
the target protein similarity network and the intermediat e layer is a drug–
target interaction bipartite graph network.
Molecular BioSystems Paper









