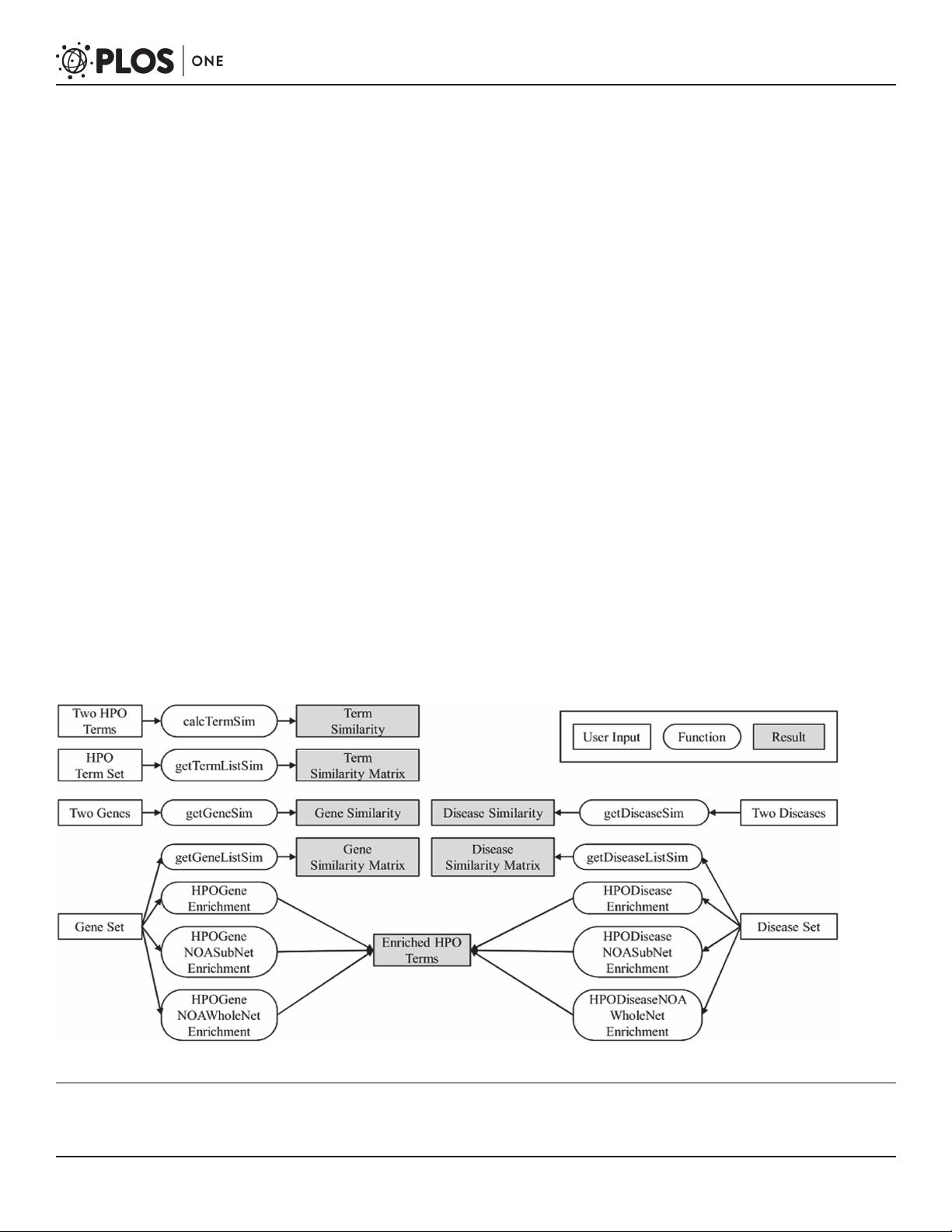
Thus, we developed an R package HPOSim with an immediate purpose to capturing pheno-
typic similarities between genes and diseases. The framework of HPOSim is shown in Fig. 1.
HPOSim analyzes semantic similarity for HPO terms, genes and diseases. Functional enrich-
ment analysis of gene set and disease set are also provided, including the classic hypergeometric
enrichment analysis and the novel network ontology analysis (NOA) [27].
Implementation
Data
HPO contains over 10000 terms (10686 terms in the HPO build #1042 released in September
2014) in three sub-ontologies, which are phenotypic abnormality (PA), onset and clinical
course (OC) and mode of inheritance (MI). Approximately 99% of the HPO terms are in the
PA sub-ontology. In each sub-ontology, terms are arranged in a directed acyclic graph (DAG)
and are related to their parent terms by “is a” relationships. The structure of the HPO allows a
term to have multiple pare nt terms, which enables different aspects of phenotypic abnormali-
ties to be explored. Diseases and genes are annotated to the most specific terms possible, which
means that if a disease or a gene is annotated to a term then all of the ancestors of this term
also apply (see Fig. 2 for an example).
The official ontology file provided by th e HPO Consortium is in obo format, which is plain
text-based. Thus, like other widely used R package for biomedical ontologies, e.g. GO.db, we
constructed an R package termed HPO.db. HPO.db provided programmatic interfaces to the
hierarchical structure of HPO terms. HPOSim uses HPO.db to obtain information about terms
and relationships between terms. HPO.db can be used by other R packages that use HPO data.
HPOSim provides two kinds of pre-calculated data within the package: the association be-
tween HPO terms, as well as association between genes and diseases (gene-to-phenotype, phe-
notype-to-gene, disease-to-phenotype and phenotype-to-disease). The associations between
HPO terms are obtained from the original ontology and annotation data provided by the HPO
Consortium, and the information content (IC) of the HPO terms is pre-calculated based on
Figure 1. Framework of HPOSim. Users can use HPOSim to calculate semantic similarity for HPO terms, genes and diseases. HPOSim can also be used
to identify enriched HPO terms for gene set and disease set.
doi:10.1371/journal.pone.0115692.g001
HPOSim: Similarity Measure and Enrichment Analysis Based on HPO
PLOS ONE | DOI:10.1371/journal.pone.0115692 February 9, 2015 3/12









