Int. J. Mol. Sci. 2018, 19, 2483 3 of 15
Int. J. Mol. Sci. 2018, 19, x FOR PEER REVIEW 3 of 15
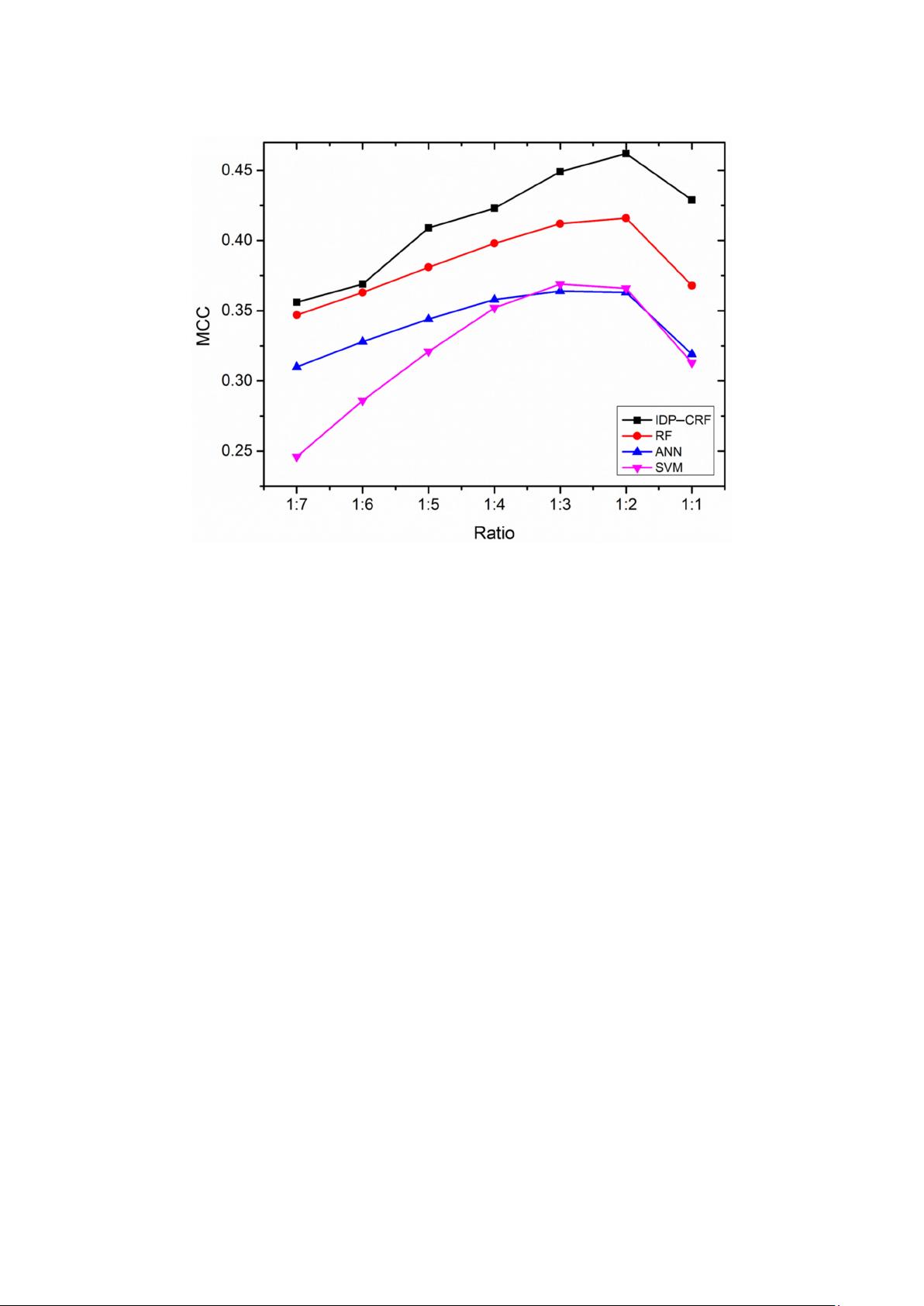
Figure 1. The performance of IDP–CRF (intrinsically disordered protein–conditional random field)
and three classification-based predictors trained with different ratios of disordered residues and
ordered residues. These three classification-based predictors include a RF (random forest) predictor,
an ANN (artificial neural network) predictor and an SVM (support vector machine) predictor. MCC
represents Matthew’s correlation coefficient performance metrics.
2.2. IDP–CRF (Intrinsically Disordered Protein–Conditional Random Field) Outperforms Classification-
Based Predictors
Sequential adjacent residues may have similar characteristics in the formation of IDPs/IDRs [18].
However, traditional classification-based predictors treat each target residue as an independent
sample, ignoring the global sequence patterns of disordered regions. To address this problem, IDP–
CRF, proposed in this study, can take the relationship between labels of sequential adjacent residues
into account. The performance of IDP–CRF and several classification-based predictors (cf. Section 3.1)
is compared by using five-fold cross-validation, and is shown in Table 1. From Table 1, we can see
that IDP–CRF obtains the highest accuracy (ACC). When the positive and negative samples are
extremely unbalanced, although ACC favors “greedy” predictions (i.e., predicting more residues as
disordered), IDP–CRF obtains the highest sensitivity (Sn) and specificity (Sp), indicating that IDP–
CRF can achieve better trade-off between Sn and Sp automatically. Besides, the highest MCC of IDP–
CRF also fully illustrates that it is an efficient predictor for identifying IDPs/IDRs. This is because
IDP–CRF can obtain more information of global sequence patterns of disordered regions compared
with classification-based predictors.
2.3. Several Examples Predicted by IDP–CRF and Three Classification-Based Predictors
In this section, three examples are used to visualize the prediction of the four predictors listed
in Table 1, including IDP–CRF, RF, SVM and ANN. These proteins are 3H2YA, 2ODKA and 4AD4A,
and their structure information is acquired from the PDB database [7]. To visualize the 3D structures
of these proteins, PyMOL [26] software is adopted to generate 3D structures of ordered regions. For
those disordered regions, their 3D structure is drawn manually.
Figure 1.
The performance of IDP–CRF (intrinsically disordered protein–conditional random field) and
three classification-based predictors trained with different ratios of disordered residues and ordered
residues. These three classification-based predictors include a RF (random forest) predictor, an ANN
(artificial neural network) predictor and an SVM (support vector machine) predictor. MCC represents
Matthew’s correlation coefficient performance metrics.
2.2. IDP–CRF (Intrinsically Disordered Protein–Conditional Random Field) Outperforms Classification-Based
Predictors
Sequential adjacent residues may have similar characteristics in the formation of IDPs/IDRs [
18
].
However, traditional classification-based predictors treat each target residue as an independent sample,
ignoring the global sequence patterns of disordered regions. To address this problem, IDP–CRF,
proposed in this study, can take the relationship between labels of sequential adjacent residues into
account. The performance of IDP–CRF and several classification-based predictors (cf. Section 3.1)
is compared by using five-fold cross-validation, and is shown in Table 1. From Table 1, we can
see that IDP–CRF obtains the highest accuracy (ACC). When the positive and negative samples are
extremely unbalanced, although ACC favors “greedy” predictions (i.e., predicting more residues
as disordered), IDP–CRF obtains the highest sensitivity (Sn) and specificity (Sp), indicating that
IDP–CRF can achieve better trade-off between Sn and Sp automatically. Besides, the highest MCC of
IDP–CRF also fully illustrates that it is an efficient predictor for identifying IDPs/IDRs. This is because
IDP–CRF can obtain more information of global sequence patterns of disordered regions compared
with classification-based predictors.
2.3. Several Examples Predicted by IDP–CRF and Three Classification-Based Predictors
In this section, three examples are used to visualize the prediction of the four predictors listed
in Table 1, including IDP–CRF, RF, SVM and ANN. These proteins are 3H2YA, 2ODKA and 4AD4A,
and their structure information is acquired from the PDB database [
7
]. To visualize the 3D structures of
these proteins, PyMOL [
26
] software is adopted to generate 3D structures of ordered regions. For those
disordered regions, their 3D structure is drawn manually.









