Articial Intelligence In Medicine 115 (2021) 102059
4
systems require expert opinion and cannot be used as a metric for model
capacity and understanding performance. Another popular explan-
ability tool SHAP (Shapely Additive Explanations) [10] describe the
contribution of each input feature towards model outcomes. However,
the lack of feature dataset in clinical decision system such as ECG
diagnosis, continuous 1D nature of ECG signals make it unsuitable for
use. None of the methods illustrated are specic to be used as metric for
understanding model capacity and performance in time series medical
datasets.
Explanations for models trained on time-series data use extracted
shapelet [15,16] (time-series subsequences) which are suited for
discovering the best patterns that are representative of a target class.
Time-series tweaking in [16] is a method applied to time-series data
although not applied to provide explanation for deep networks.
Time-series tweaking nds the minimum number of changes needed in
order to change an input classication outcome in a random forest type
of classier. These time series explanations cannot be used as metric for
evaluation of model performance. While CEFEs does not use shapelets to
extract model learned features, it uses ECG waveform segmentation
techniques to discover, map and compare model learned features to
those in input ECG signals. The methods in [9–11] and [27–29] focused
input feature scoring and data perturbation differ with our proposed
CEFEs which provides interpretable insights on specic features learned
by a 1D-CNN model and explanations on how these learned features
affect CNN model capacity and outcomes.
In summary, literature survey shows majority of interpretation and
explanations research has been on 2D-CNN models in non-medical
domain, leaving a gap of explanation of medical time-series data.
Traditional metrics such as accuracy, sensitivity, and selectivity are not
sufcient for providing details of structural ECG features learned by a
CNN model. The challenges posed by medical signal datasets as dis-
cussed in the Introduction section, hinders ability of CNN to learn,
especially specic intricate structural clinical features for clinical diag-
nosis. Developing interpretable and explainable techniques for health-
care timeseries data creates supportive trust and condence in
automated decision support systems. CEFEs framework addresses these
gaps by providing interpretable explanations for CNN models trained on
ECG timeseries data, by focusing on post-hoc model interpretability in
terms of model capacity.
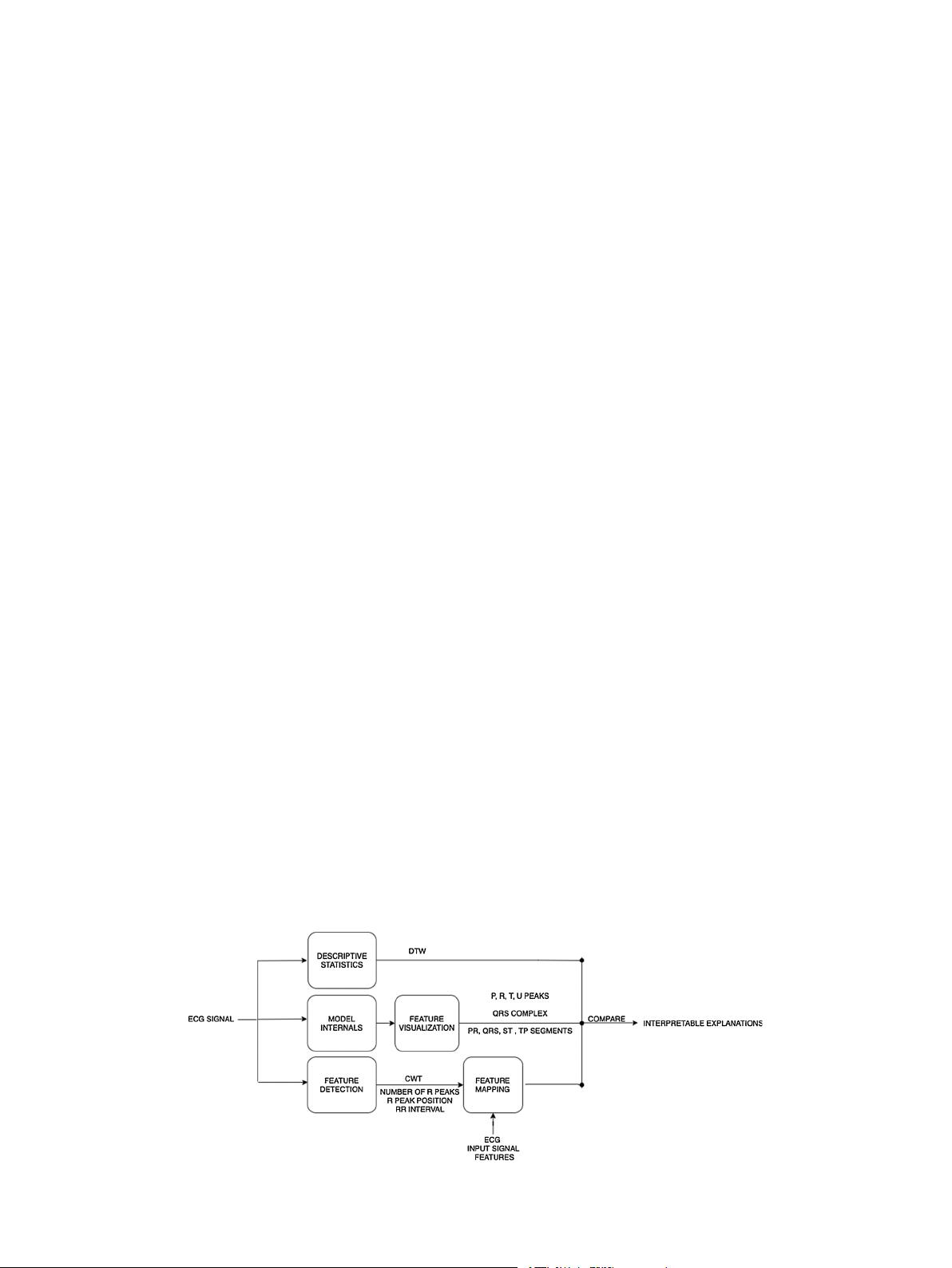
3. CEFEs
We aim to provide transparency and functional understanding of 1D-
CNN model using a layer-wise interpretation of relevant features learned
by the model. Denitions of Interpretation and Explanation in the context
of computation models are often used interchangeably. Montavon et al.
[1] denes Interpretation as the idea of mapping from feature space (e.g.,
predicted class) into a human comprehendible domain and Explanations
as a set of features in the interpretable domain that contribute towards
class discrimination.
Our proposed framework (Figs. 2 and 3) for ECG signals, is a post-hoc
tri-modular evaluation structure that provides local interpretations and
explanations from convolution neural networks. Local interpretations
and explanations of a model explain the “why” of individual test case
predictions. In this section, we present the details of CEFEs modules and
the process by which the framework achieves model interpretation and
explanations.
1 Descriptive Statistics: Descriptive statistics are summary analysis of
representative model features or input data. These representations
help users realize a model’s capacity to learn inherent statistical and
mechanical features of data such as waveform shape features of
signal. CEFEs descriptive statistics module uses task dependent tests
to analyze an input ECG signal and corresponding feature map
extracted from a convolution layer of a trained CNN model. Although
the choice of CNN layer for statistical analysis is not limited to a
specic layer, we were motivated to use the nal convolution layer
(Conv
nal
) because this layer incorporates both low level and high-
level data features and balances spatial and semantics information
contribute to explainable and interpretable class discrimination ar-
tifacts. Descriptive statistics tests are task dependent. We chose Dy-
namic Time Warping (DTW) algorithm to compute the similarities
between the input ECG signal and the CNN model learned features.
DTW enabled us to analyze and observe learned representation of the
rigid ECG signal morphology. DTW distance measures are organized
into intra-model distance (Eq. 1) and inter-model distance (Eq. 2).
Intra-model distance (d
intra
) is the warped Euclidean similarity
measure returned by DTW from an input ECG signal and feature map
projections. We dene (d
intra
) as a value that represents how well a
model has learned input ECG shape features. A low (d
intra
) value
explains that a model has adequately learned ECG shape features.
Once (d
intra
) values of several models are computed, we compute the
difference in learned ECG shape features between two CNN models
using the inter-model distance (d
inter
). The (d
inter
) values are used as a
comparative measure of ECG shape features learned between two
models trained on similar input ECG signals. A high (d
inter
) value
explains the differences in prediction outcomes of two models on a
xed test set [17].
d
intra
=
K
k=1
x
k,m
− y
k,n
∗
x
k,m
− y
k,n
(1)
d
inter
= |d
M
y1
intra
− d
M
y2
intra
| (2)
Where k represents the samples, m
th
data point of one input signal (ECG
Signal), n
th
data point of other input signal (Feature Map) and M
y1
, M
y2
represent the two models under comparison. We approximate d
inter
and
Fig. 3. CEFEs - Explainable Modules.
B.M. Maweu et al.









