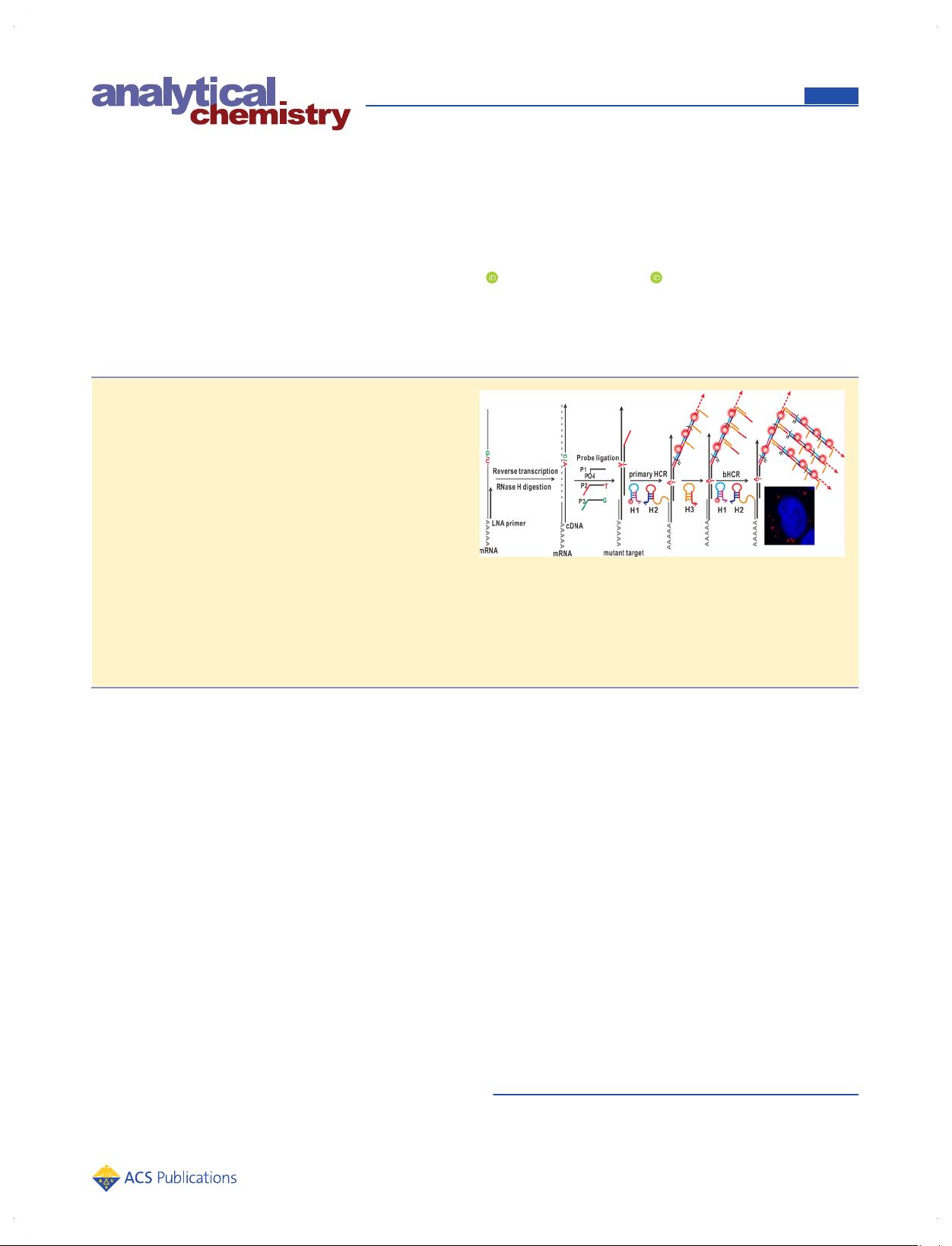
In Situ Imaging of Individual mRNA Mutation in Single Cells Using
Ligation-Mediated Branched Hybridization Chain Reaction (Ligation-
bHCR)
Ying Tang, Xiao-Li Zhang, Li-Juan Tang, Ru-Qin Yu, and Jian-Hui Jiang*
State Key Laboratory of Chemeo/Bio-Sensing and Chemometrics, Institute of Chemical Biology and Nanomedicine, College of
Chemistry and Chemical Engineering, Hunan University, Changsha 410082, P. R. China
*
S
Supporting Information
ABSTRACT: Ultrasensitive and specific in situ imaging of
gene expression is essential for molecular medicine and clinical
theranostics. We develop a novel fluorescence in situ
hybridization (FISH) strategy based on a new branched
hybridization c hain reaction (bHCR) for efficient signal
amplification in the FISH assay and a ligase-mediated
discrimination for specific mutation detection. To our
knowledge, this is the first time that HCR has been realized
for mutation detection in the FISH assay. In vitro assay shows
that the ligation-bHCR strategy affords high specificity in
discriminating single-nucleotide variation in mRNA, and it
generates a highly branched polymeric product that confers more efficient amplification or better sensitivity than HCR. Imaging
analysis reveals that ligation-bHCR generates highly bright spot-like signals for localization of individual mRNA molecules, and
spot signals of different colors are highly specific in genotyping point mutation of individual mRNA. Moreover, this strategy is
shown to have the potential for quantitative imaging of the expression of mRNA at the single-cell level. Therefore, this strategy
may provide a new promising paradigm in developing highly sensitive and specific FISH methods for various diagnostic and
research applications.
D
etection of gene expression in signal transduction
pathways is essential for be tter unde rstanding of
mechanisms underlying various diseases and for development
of precise theranostic design s.
1
Curr ent population-based
assays, which are performed on crude tissue extracts to analyze
the content from many cells, typically fail to provide gene
signatures in rare cells.
2
Increasing knowledge about cellular
and tissue heterogeneity in complex diseases such as cancer has
highlighted the demand for gene detection technologies at the
single-cell level, preferentially with single-molecule sensitivity.
3,4
Fluorescence in situ hybridization (FISH) is a pivotal tool for
genetic analysis at single-cell resolution, which allows direct
mapping of gene expression in a morphological context in the
subcellular and tissue scale.
5
However, current methods of
FISH still suffers from limitations of low signal intensity or
sensitivity and compromised specificity.
5
These limitations
create hurdles for FISH in meeting the needs critical for various
diagnostic and research applications, such as detecting genes
such as mRNA with very low copy numbers and resolving a
sequence of high similarity such as point mutations.
6
Development of the next-generation FISH methodologies
that enable ultrasensitive and highly selective in situ imaging
of mRNA point mutation at single-molecule resolution remains
a great challenge.
Rolling-circle amplification (RCA)-based
7,8
FISH (RCA-
FISH) is a useful strategy for highly sensitive and selective
genetic analysis of DNA,
9
mRNA,
10
and miRNA.
11,12
The
RCA-FISH strategy is able to provide information about the
localization of nucleic acid targets at the single-molecule level
with single-molecule sensitivity. Recent effort in RCA assay has
been successfully implemented for in situ detection of mRNA
point mutations
13
and protein complexes.
14
Despite the
success, RCA-FISH typically shows a limited detection
efficiency because in situ ligation of circular probes may be
subjected to topological constraints and lower efficiency than
that for linear probes, and amplification reaction may be
blocked by nucleic acid targets or their binding proteins.
15,16
To
address this difficulty, hybridization chain reaction (HCR), an
emerging nonenzymatic nucleic acid circuits for signal
amplification and detection,
17,18
has been recently developed
for multiplexed FISH of mRNA.
19
Without the need for
polymerase-based amplification, HCR-F ISH exh ibit s deep
penetration, high contrast, and sharp localization. However,
because stringent hybridization conditions for FISH reduce the
energetic driving force for HCR polymerization and thus
decrease the sensitivity, complicated optimization of HCR
probes has to used to increase the signal-to-background ratio in
Received: November 3, 2016
Accepted: February 20, 2017
Published: February 20, 2017
Article
pubs.acs.org/ac
© 2017 American Chemical Society 3445 DOI: 10.1021/acs.analchem.6b04312
Anal. Chem. 2017, 89, 3445−3451









