generations, is satisfied, the non-dominated antibodies in external
archive are exported as the final result.
When initialization, an initial population P ¼fx
1
; x
2
; :::; x
N
g is
created, where the decision variables of x
i
ði ¼ 1; 2; :::; NÞ are ran-
domly generated in Ω and the corresponding objective values are
evaluated. After performing the fast non-dominated sorting [16] in
P, the non-dominated and dominated individuals are obtained and
respectively preserved in the external archive (EXA) and domi-
nated archive (DA). The pseudo-code of initialization, named
Algorithm 1, is described in Fig. 2.
3.1. Cloning operator
It is assumed that the evolutionary population is P with size N
and the cloning population is P
C
¼fa
1
; a
2
; :::; a
NC
g with size NC.At
first, NC antibodies having the larger crowding-distance values are
selected from EXA. Afterward, the cloning is activated and the
evolutionary population will be
P ¼[
NC
i ¼ 1
fq
i
a
i
gð6Þ
where q
i
stands for the number of clones corresponding to each
antibody a
i
ði ¼ 1; 2; :::; NCÞ, calculated by
q
i
¼ N
CDða
i
Þ
P
NC
j ¼ 1
CDða
j
Þ
&’
ð7Þ
where CDða
i
Þ is the fitness value of antibody a
i
ði ¼ 1; 2; :::; NC Þ and
set as the crowding-distance value, computed by the following
equation:
CDða
i
Þ¼
X
m
j ¼ 1
CD
j
ða
i
Þ
f
j
max f
j
min
ð8Þ
where m is the number of objectives, f
j
max and f
j
min are
respectively the maximal and minimal values of the jth objective,
CD
j
ða
i
Þ is the crowding distance of the jth objective for the
antibody a
i
ði ¼ 1; 2; :::; NC Þ and obtained by
It is noted that when the antibody locates in the boundary, its
crowding distance is 1 and the number of clones cannot be
obtained by Eq. (7). In this case, the crowding distance of the
boundary antibody is set as twice of the maximal crowding
distance except for the boundary solutions. The pseudo-code of
this operator named Algorithm 2 is shown in Fig. 3, where NC is
the size of clone population and the function CrowdingDistan-
ceAssignment(P) calculates the crowding distance value of each
antibody in population P.
3.2. Evolutionary operator
3.2.1. DE operator
Differential evolution has very strong global search capability
and shows pretty good convergence ability [27–34]. Thus, DE
operator is widely adopted in MOEAs [29–34]. In ADE-MOIA, a
novel ADE operator is presented, which is tailor-made to effec-
tively cooperate with MOIA. The weakness of MOIAs in population
diversity can be repaired by the strong global search capability of
DE. Our proposed ADE operator includes a suitable parent selec-
tion strategy and a novel adaptive parameter control approach.
This makes ADE operator become more effective for MOIAs. The
ADE operation is given as follows:
v
i
¼ x
r
1
þFðx
r
2
x
r
3
Þð10Þ
where F A 0ð ; 1:0
is a scaling factor. It is important to note that the
selected parent vectors used in Eq. (10) have great impact on the
optimization performance. The most popular DE operators include
DE/rand/1/bin, DE/best/1/bin, DE/current-to-best/1/bin, DE/rand/
2/bin, DE/best/2/bin [28], which are mostly adopted to solve SOPs
and especially effective for some SOPs with certain complex
features. When solving MOPs, these popular DE operators are
not perfectly fit as all the non-dominated solutions are regarded as
the best candidate for MOPs. The selection of parent vectors in
MOPs is challenging. In our proposed ADE operator, in order to
promote the population diversity, three parent vectors picked
from the EXA and DA are aimed at providing a correct evolutionary
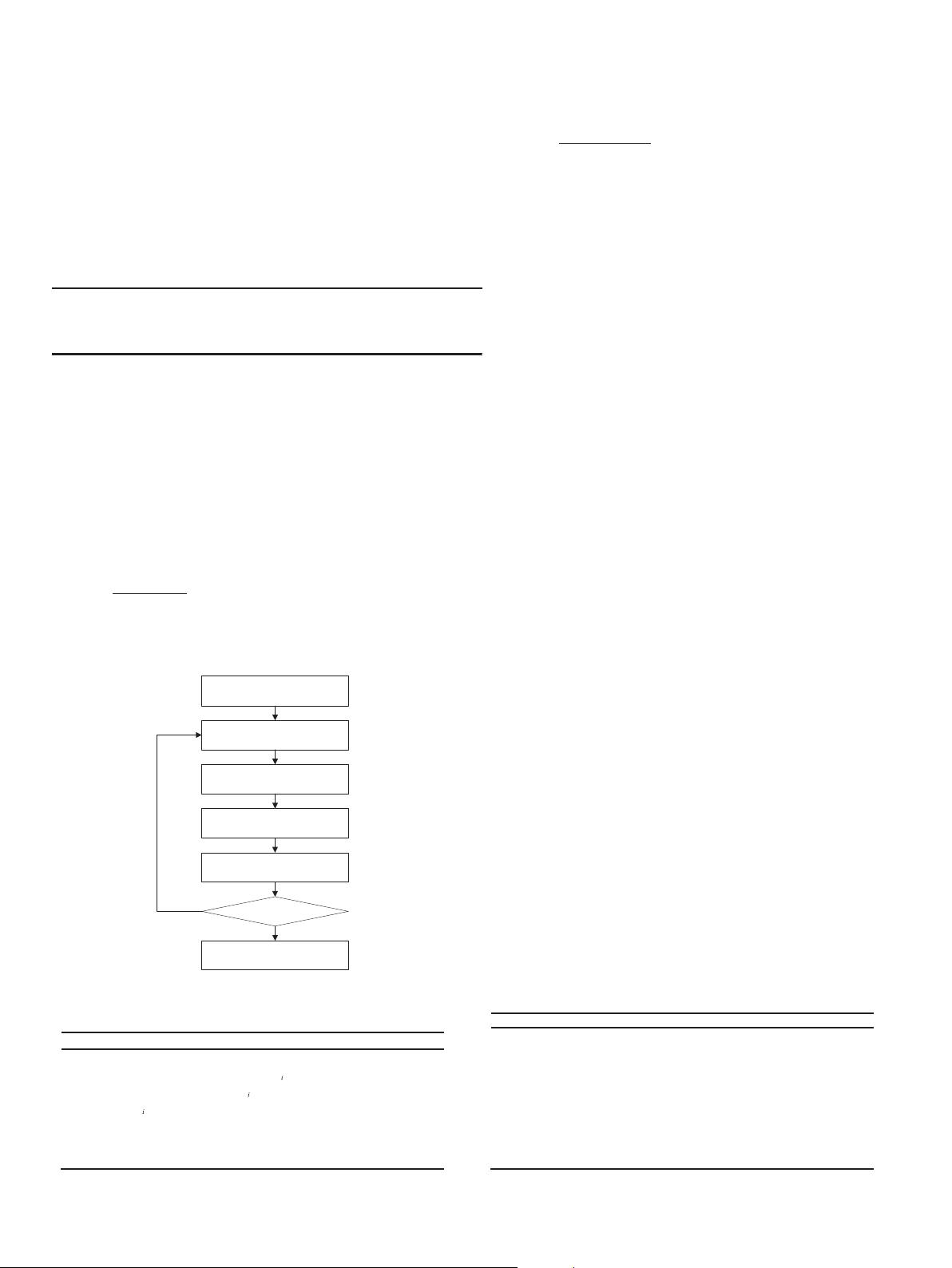
ADE operator
Update archive
Cloning operator
Initialization
Output result
Yes
No
Perturbance
Termination?
Fig. 1. The algorithmic flowchart of ADE-MOIA.
Algorithm 1: Initialization
randomly generate an individual
evaluate the objectives of
EXA = Find_nonDominated( P ); //find non-dominated solutions in P
DA = Find_Dominated ( P ); //find the dominated solutions in P
Fig. 2. The pseudo-code of initialization.
Algorithm 2: Cloning Operator
CrowdingDistanceAssignment(P
C
);
P
C
= Sort( P
C
);// sort P
C
according to the crowding distance in descending order
P
C
= SelectforClone( P
C
);//select the first NC antibodies in P
C
calculate the number q
i
of clones for
i
x
Fig. 3. The pseudo-code of cloning operator.
CD
j
ða
i
Þ¼
1; if ðf
j
ða
i
Þ¼¼f
j
min or f
j
ða
i
Þ¼¼f
j
maxÞ;
min ff
j
ða
k
Þf
j
ða
l
Þg; f
j
ða
k
Þ4 f
j
ða
i
Þ4 f
j
ða
l
Þðk; lA ½1; NÞ; otherwise
(
ð9Þ
Q. Lin et al. / Computers & Operations Research 62 (2015) 95–11198









