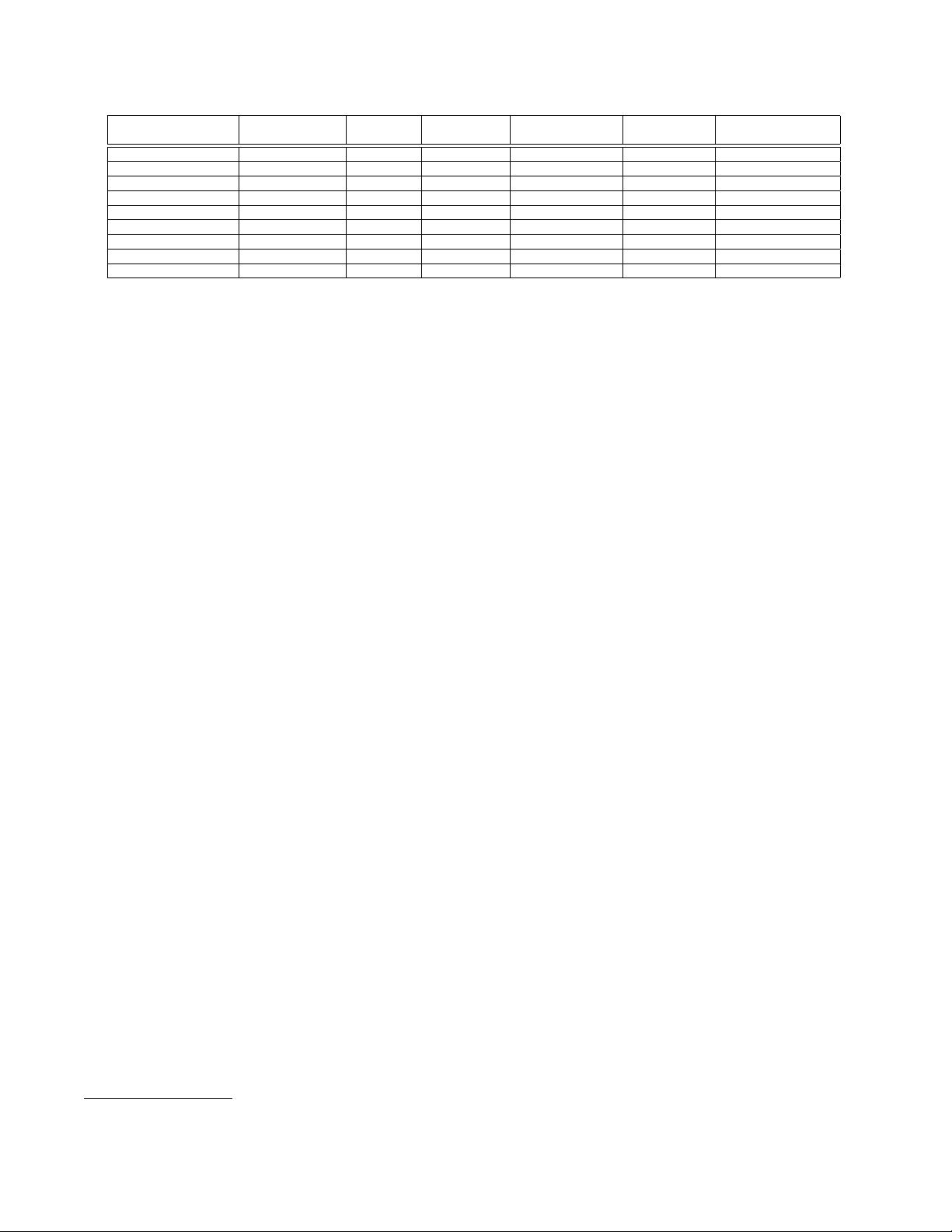
Table 1: Overview of parallel density-based clustering algorithms.
Name Parallel model
Implements
DBSCAN
Approximated Partitioner required
Data object
type
Distance function
supported
ρ-DBSCAN [12] single machine yes yes data on a grid point in n-D Euclidean
MR-DBSCAN [14] MapReduce yes no yes point in n-D Euclidean
SPARK-DBSCAN Apache Spark yes no yes point in n-D Euclidean
IRVINGC-DBSCAN Apache Spark yes no yes point in 2-D Euclidean
DBSCAN-MR [6] MapReduce yes no yes point in n-D Euclidean
MR. SCAN [30] MRNet + GPGPU yes no yes point in 2-D Euclidean
PARDICLE [25] MPI yes yes yes point in n-D Euclidean
DBCURE-MR [16] MapReduce no no yes point in n-D Euclidean
NG-DBSCAN MapReduce yes yes no arbitrary type arbitrary symmetric
which consider the input graph as given, is that our approach builds
the graph in its first phase; doing this efficiently is not trivial, since
some of the most common choices (such as ε-neighbor or k-nearest
neighbor graphs) require O(n
2
) computational cost for generic dis-
tance functions; our approximated approach obtains a substantial
cut on these costs.
2.4 Density-Based Clustering
for High-Dimensional Data
We conclude our discussion of related work with density-based
approaches suitable for text and high-dimensional data in general.
Tran et al. [28] propose a method to identify clusters with dif-
ferent densities. Instead of defining a threshold for a local density
function, low-density regions separating two clusters can be de-
tected by calculating the number of shared neighbors. If the num-
ber of shared neighbors is below a threshold, then the two objects
belong to two different clusters. Tran et al. report that their ap-
proach has high computational complexity, and the algorithm was
evaluated using only a small dataset (below 1 000 objects). In addi-
tion, as the authors point out, this approach is unsuited for finding
clusters that are very elongated or have particular shapes.
Zhou et al. [32] define a different way to identify dense regions.
For each object p, their algorithm computes the ratio between the
size of p’s ε-neighborhood and those of its neighbors, to distinguish
nodes that are at the center of clusters. This approach is once again
only evaluated and compared with DBSCAN in a 2D space.
3. NG-DBSCAN: APPROXIMATE AND
FLEXIBLE DBSCAN
NG-DBSCAN is an approximate, distributed, scalable algorithm
for density-based clustering, supporting any symmetric distance
function. We adopt the vertex-centric, or “think like a vertex” pro-
gramming paradigm, in which computation is partitioned by and
logically performed at the vertexes of a graph, and vertexes ex-
change messages. The vertex-centric approach is widely used due
to its scalability properties and expressivity [23].
Several vertex-centric computing frameworks exist [1, 13, 22]:
these are distributed systems that iteratively execute a user-defined
program over vertices of a graph, accepting input data from adja-
cent vertices and emitting output data that is communicated along
outgoing edges. In particular, our work relies on frameworks sup-
porting Valiant’s Bulk Synchronous Parallel (BSP) model [29], which
employs a shared nothing architecture geared toward synchronous
execution. Next, for clarity and generality of exposition, we gloss
over the technicalities of the framework, focusing instead on the
principles underlying our algorithm. Our implementation uses the
Apache Spark framework; its source code is available online.
1
1
https://github.com/alessandrolulli/gdbscan
3.1 Overview
Together with efficiency, the main design goal of NG-DBSCAN
is flexibility: indeed, it handles arbitrary data and distance func-
tions. We require that the distance function d is symmetric: that is,
d(x, y) = d(y, x) for all x and y. It would be technically possi-
ble to modify NG-DBSCAN to allow asymmetric distance, but for
clustering – where the goal is grouping similar items – asymmetry
is conceptually problematic, since it is difficult to choose whether
x should be grouped with y if, for example, d(x, y) is large and
d(y, x) is small. If needed, we advise using standard symmetriza-
tion techniques: for example, defining a d
0
(x, y) equal to the mini-
mum, maximum or average between d(x, y) and d(y, x) [11].
The main reason why DBSCAN is expensive when applied to
arbitrary distance measures is that it requires retrieving each point’s
ε-neighborhood, for which the distance between all node pairs needs
to be computed, resulting in O
n
2
calls to the distance function.
NG-DBSCAN avoids this cost by dropping the requirement of
computing ε-neighborhoods exactly, and proceeds in two phases.
The first phase creates the ε-graph, a data structure which will
be used to avoid ε-neighborhood queries: ε-graph nodes are data
points, and each node’s neighbors are a subset of its ε-neighbor-
hood. This phase is implemented through an auxiliary graph called
neighbor graph which gradually converges from a random starting
configuration towards an approximation of a k-nearest neighbor (k-
NN) graph by computing the distance of nodes at a 2-hop distance
in the neighbor graph; as soon as pairs of nodes at distance ε or less
are found, they are inserted in the ε-graph.
The second phase takes the ε-graph as an input and computes
the clusters which are the final output of NG-DBSCAN; cheap
neighbor lookups on the ε-graph replace expensive ε-neighborhood
queries. In its original description, DBSCAN is a sequential algo-
rithm. We base our parallel implementation on the realization that
a set of density-reachable core nodes corresponds to a connected
component in the ε-graph– the graph where each core node is con-
nected to all core nodes in its ε-neighborhood. As such, our Phase
2 implementation builds on a distributed algorithm to compute con-
nected components, amending it to distinguish between core nodes
(which generate clusters), noise points (which do not participate to
this phase) and border nodes (which are treated as a special case,
as they do not generate clusters).
NG-DBSCAN’s parameters determine a trade-off between speed
and accuracy, in terms of fidelity of the results to the exact DB-
SCAN implementation: in the following, we describe in detail our
algorithm and its parameters; in Section 5.4, we quantify this trade-
off and provide recommended settings.
3.2 Phase 1: Building the ε-Graph
As introduced above, Phase 1 builds the ε-graph, that will be
used to avoid expensive ε-neighborhood queries in Phase 2. We
159









