Today
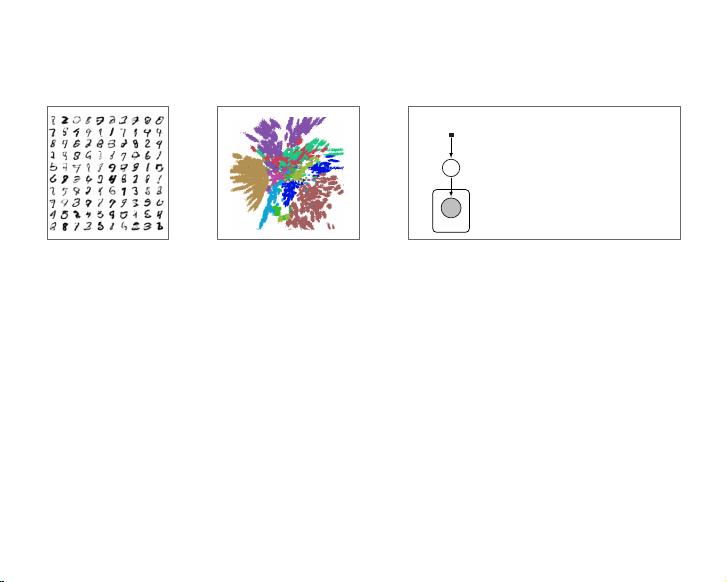
(a) Learned Frey Face manifold (b) Learned MNIST manifold
Figure 4: Visualisations of learned data manifold for generative models with two-dimensional latent
space, learned with AEVB. Since the prior of the latent space is Gaussian, linearly spaced coor-
dinates on the unit square were transformed through the inverse CDF of the Gaussian to produce
values of the latent variables z. For each of these values z, we plotted the corresponding generative
p
✓
(x|z) with the learned parameters ✓.
(a) 2-D latent space (b) 5-D latent space (c) 10-D latent space (d) 20-D latent space
Figure 5: Random samples from learned generative models of MNIST for different dimensionalities
of latent space.
B Solution of D
KL
(q
(z)||p
✓
(z)), Gaussian case
The variational lower bound (the objective to be maximized) contains a KL term that can often be
integrated analytically. Here we give the solution when both the prior p
✓
(z)=N(0, I) and the
posterior approximation q
(z|x
(i)
) are Gaussian. Let J be the dimensionality of z. Let µ and
denote the variational mean and s.d. evaluated at datapoint i, and let µ
j
and
j
simply denote the
j-th element of these vectors. Then:
Z
q
✓
(z) log p(z) dz =
Z
N(z; µ,
2
) log N(z; 0, I) dz
=
J
2
log(2⇡)
1
2
J
X
j=1
(µ
2
j
+
2
j
)
10
Stochastic Back-propagation in DLGMs
(a) NORB (b) CIFAR (c) Frey
Figure 4. a) Performance on the NORB dataset. Left: S a mp l es from the training data. Right: sampled pixel means from
the model. b) Performance on CIFAR10 patches. Left: Samples from the training data. Right: Sampled pixel means
from the model. c) Frey faces data. Left: data samples. Right: model samples.
Figure 5. Imputation results on MNIST digits. The first
column shows the true data. Column 2 shows pixel loca-
tions set as missing in grey. The remaining columns show
imputations a n d denoising of the i ma g es for 15 iterations,
starting left to right. Top: 60% missingness. Middle: 80%
missingness. Bottom: 5x5 patch missing.
matics and experimental design. We show the ability
of the model to impute missing data using the MNIST
data set in figure 5. We test the imputation ability
under two di↵erent missingness types (Little & Rubin,
1987): Missing-at-random (MAR), where we consider
60% and 80% of the pixels to be missing randomly, and
Not Missing-at-random (NMAR), where we consider a
square region of the image to be missing. The model
produces very good completions in both test cases.
There is uncertainty in the identity of the image. This
is expected and reflected in the errors in these comple-
tions as the resampling procedure is run, and furth er
demonstrates the ability of the model to capture the
diversity of the und er l yi n g data. We do not integrate
over the missing values in our imputation procedure,
but use a procedure that simulates a Markov chain
that we show converges to the true marginal distribu-
tion. The procedure to sample from the missing pixels
given the observed pixels is explained in appendix E.
Figure 6. Two dimensional embedding of the MNIST data
set. Each colour correspo n d s to one of the digit classes.
6.5. Data Visualisation
Latent variable models such as DLGMs are often used
for visualisation of high-dimensional data sets. We
project the MNIST data set to a 2-dimensional latent
space and use this 2-D embedding as a visualisation of
the data. A 2-dimensional embedding of the MNIST
data set is shown in figure 6. The classes separate
into di↵erent regions indicating that such a tool can
be useful in gaining insight into the structure of high-
dimensional data sets.
7. Discussion
Our algorithm generalises to a large class of models
with continuous latent variables, which include Gaus-
sian, non-negative or sparsity-promoting latent vari-
ables. For models with discrete latent variables (e.g.,
sigmoid belief networks), policy-gradient approaches
that improve upon the REINFORCE approach remain
the most general, but intelligent design is needed to
control t h e gradient-variance in high dimensional set-
tings.
These models are typically used with a large number
[Kingma and Welling 2013] [Rezende et al. 2014]
x
n
✓
˛ D 1:5; D 1
N
data {
int N; // number of observations
int x[N]; // discrete - valued observations
}
parameters {
// latent variable , must be positive
real < lower=0> th eta ;
}
model {
// non - conjugate prior for latent variable
theta ~ weibul l (1.5 , 1) ;
// likelihood
for (n in 1:N)
x[n] ~ poisson(theta);
}
Figure 2: Specifying a simple nonconjugate probability model in Stan.
analysis posits a prior density p.✓/ on the latent variables. Combining the likelihood with the prior
gives the joint density p.X; ✓/ D p.X j ✓/p.✓/.
We focus on approximate inference for differentiable probability models. These models have contin-
uous latent variables ✓. They also have a gradient of the log-joint with respect to the latent variables
r
✓
logp.X; ✓/. The gradient is valid within the support of the prior supp.p.✓ // D
˚
✓ j ✓ 2
R
K
and p.✓/>0
✓ R
K
, where K is the dimension of the latent variable space. This support set
is important: it determines the support of the posterior density and plays a key role later in the paper.
We make no assumptions about conjugacy, either full or conditional.
2
For example, consider a model that contains a Poisson likelihood with unknown rate, p.x j ✓ /. The
observed variable x is discrete; the latent rate ✓ is continuous and positive. Place a Weibull prior
on ✓, defined over the positive real numbers. The resulting joint density describes a nonconjugate
differentiable probability model. (SeeFigure 2.) Its partial derivative@=@✓ p.x; ✓ / is valid within the
support of the Weibull distribution, supp.p.✓ // D R
C
⇢ R. Because this model is nonconjugate, the
posterior is not a Weibull distribution. This presents a challenge for classical variational inference.
In Section 2.3, we will see how handles this model.
Manymachinelearning models are differentiable. Forexample: linear and logisticregression, matrix
factorization with continuous or discrete measurements, linear dynamical systems, and Gaussian pro-
cesses. Mixture models, hidden Markov models, and topic models have discrete random variables.
Marginalizing out these discrete variables renders these models differentiable. (We show an example
in Section 3.3.) However, marginalization is not tractable for all models, such as the Ising model,
sigmoid belief networks, and (untruncated) Bayesian nonparametric models.
2.2 Variational Inference
Bayesian inference requires the posterior density p.✓ j X/, which describes how the latent variables
vary when conditioned on a set of obser vations X. Many posterior densities are intractable because
their normalization constants lack closed forms. Thus, we seek to approximate the posterior.
Consider an approximating density q.✓ I / parameterized by . We make no assumptions about its
shape or support. We want to find the parameters of q.✓ I / to best match the posterior according to
some loss function. Variational inference () minimizes the Kullback-Leibler () divergence from
the approximation to the posterior [2],
⇤
D argmin
KL
.
q.✓ I / k p.✓ j X/
/
: (1)
Typically the divergencealso lacks a closed form. Instead we maximize the evidence lower bound
(), a proxy to the divergence,
L./ D E
q.✓/
⇥
logp.X; ✓/
⇤
E
q.✓/
⇥
logq.✓ I /
⇤
:
The first term is an expectation of the joint density under the approximation, and the second is the
entropy of the variational density. Maximizing the minimizes the divergence [1, 16].
2
The posterior of a fully conjugate model is in the same family as the prior; a conditionally conjugate model
has this property within the complete conditionals of the model [3].
3
There is now a flurry of new work on variational inference, making it
scalable, easier to derive, faster, more accurate, and applying it to more
complicated models and applications.
Modern VI touches many important areas: probabilistic programming,
reinforcement learning, neural networks, convex optimization, Bayesian
statistics, and myriad applications.
Our goal today is to teach you the basics, explain some of the newer ideas,
and to suggest open areas of new research.