Nellore et al. Genome Biology
(2016) 17:266
Page 3 of 14
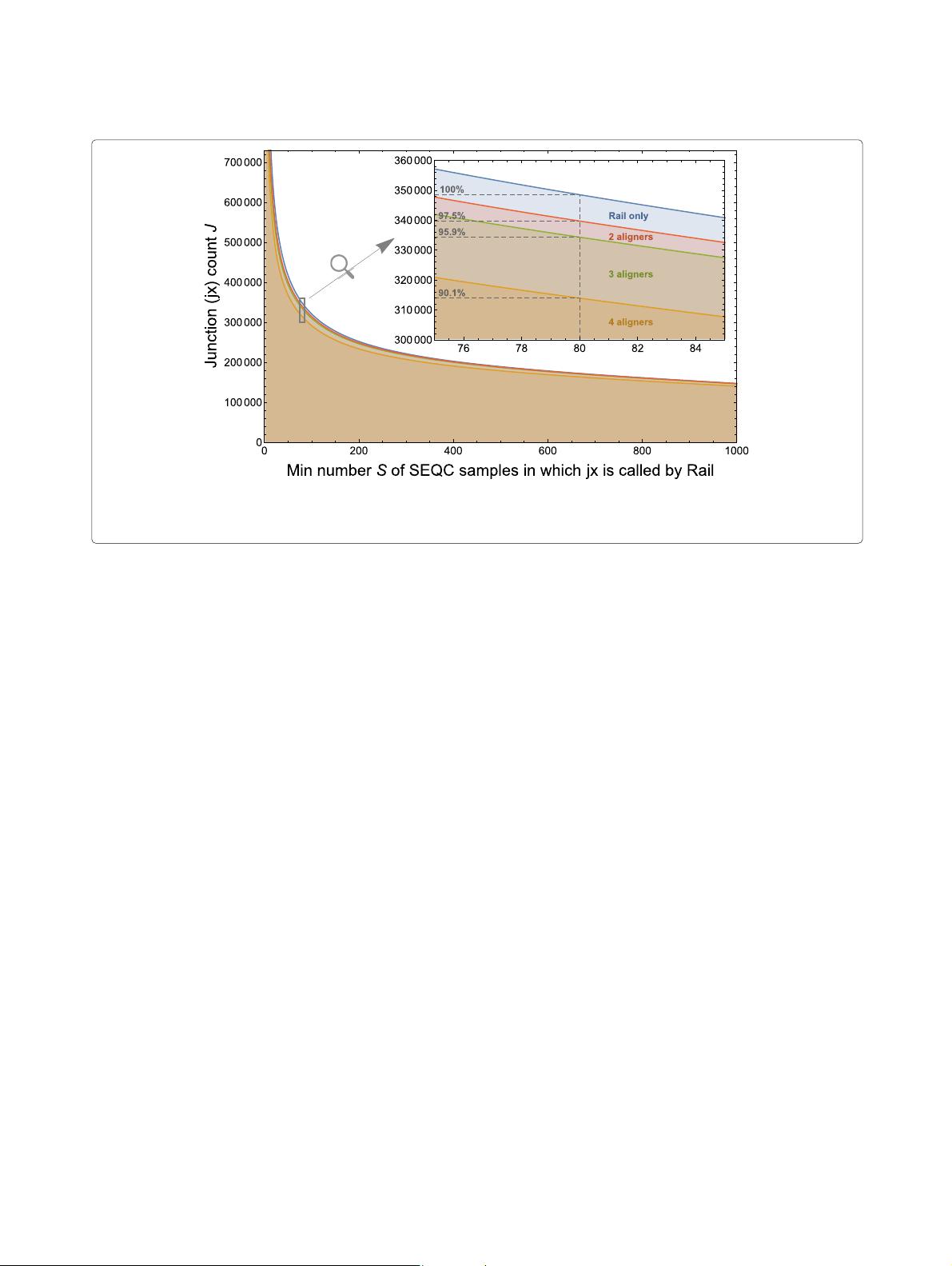
Fig. 1 Displayed is the number of exon-exon junctions J found by Rail-RNA and other alignment protocols in at least S of the 1720 brain and universal
human reference RNA-seq samples also studied by the SEQC/MACQ-III consortium [11] (i.e., SEQC). “2 aligners” (red), “3 aligners” (green), and “4
aligners” (orange) refer to junctions we found with Rail-RNA that were also found by, respectively, 1, 2, and 3 of the alignment protocols used by SEQC
of cutoffs. For each RNA-seq junction we considered, we
also evaluated whether it appeared in annotation. We con-
sidered the following levels of evidence: (1) fully annotated
junctions; (2) separately annotated junctions (typically
exon-skipping events), where both the donor and accep-
tor sites appear in one or more junctions from annotation,
but never for the same junction; (3) alternative donor and
acceptor sites, where only either the donor or the accep-
tor site appears in one or more junctions from annotation;
and (4) novel junctions, where neither donor nor acceptor
site is found in any annotated junction.
We observed that the RNA-seq junctions most widely
expressed across samples and experiments were well doc-
umented in annotation. For example, we observed that
junctions that appeared in at least 40% of human RNA-
seq samples on the SRA (S ≥ 8000) were also present in
previous annotation at least 99.8% of the time. However,
18.6% of junctions that appeared in 1000 or more sam-
ples did not appear in annotation (Fig. 2a). Many of these
unannotated junctions are partially annotated, but 3.5% of
junctions found in more than 1000 samples do not match
any splice site from an annotated junction.
We also took an investigator-focused view of the rela-
tionship between annotation and expression. Most inves-
tigators collect only a small number of samples for their
study. We restricted attention to samples where at least
100,000 RNA-seq junctions were found to rule out obvi-
ously small RNA-seq samples and samples that were
mislabeled as RNA-seq on the SRA. In each sample, we
counted the number of instances where a read maps
across a junction. (A read mapping across two junctions
thus contributes two instances.) The total number of such
“junction overlaps” across samples is a measure of the
total expression of junctions across the SRA. Most of
the reads that map to junctions map to annotated junc-
tions (Fig. 2b). In 10,090 of a total of 10,311 samples that
meet our criterion of 100,000 junctions observed, more
than 95% of junction overlaps correspond to annotated
junctions.
This represents only the bulk coverage of junctions.
We can also consider the number of junctions observed,
regardless of coverage. In 3389 out of 10,311 samples,
we observe that fewer than 80% of junctions appear in
annotation (Fig. 2c). So while the most highly covered
junctions are well annotated, there is a large number
of junctions that are well covered across multiple sam-
ples but may not appear in any given small subset of
samples.
To explore this idea further, we investigated the poten-
tial for single studies to be the sole contributors of individ-
ual unannotated junctions. In this event, the junction may
not have been called robustly across experimental proto-
cols. Here, we considered junctions that appeared in at
least P projects instead of samples. We again broke this
calculation down by the different potential levels of evi-
dence: whether the junction was entirely novel, had an
alternative donor or acceptor, an exon skip, or whether
it was fully annotated (Fig. 3). The story at the project
level mirrors the story at the sample level: 23.4% of junc-
tions found in more than 200 of the 929 projects are not
fully annotated. So unannotated junctions recur across
independent investigations.

















