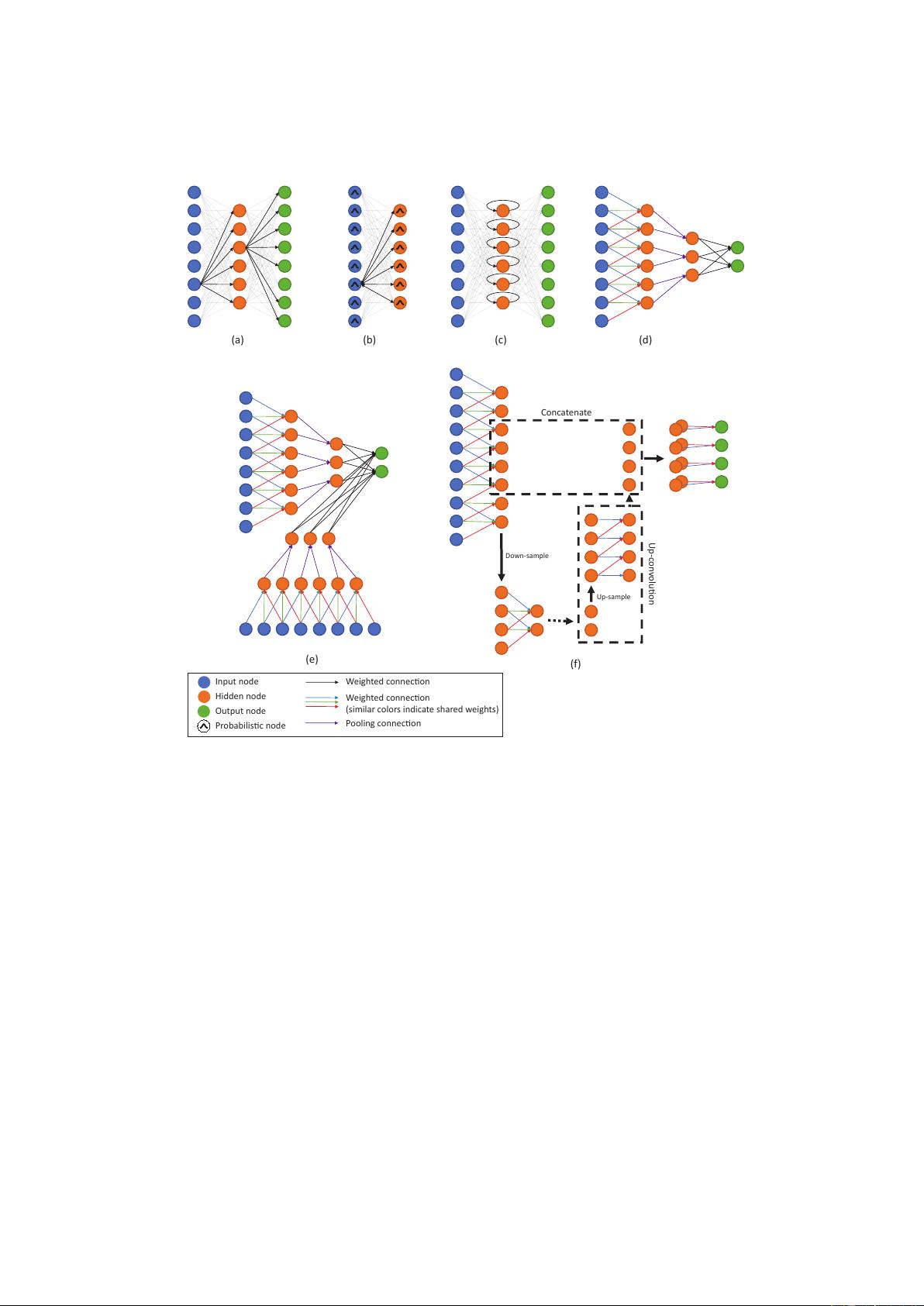
2.6.2. Restricted Boltzmann Machines (RBMs) and
Deep Belief Networks (DBNs)
RBMs (Hinton, 2010) are a type of Markov Ran-
dom Field (MRF), constituting an input layer or visi-
ble layer x = (x
1
, x
2
, . . . , x
N
) and a hidden layer h =
(h
1
, h
2
, . . . , h
M
) that carries the latent feature represen-
tation. The connections between the nodes are bi-
directional, so given an input vector x one can obtain
the latent feature representation h and also vice versa.
As such, the RBM is a generative model, and we can
sample from it and generate new data points. In anal-
ogy to physical systems, an energy function is defined
for a particular state (x, h) of input and hidden units:
E(x, h) = h
T
Wx − c
T
x − b
T
h, (9)
with c and b bias terms. The probability of the ‘state’ of
the system is defined by passing the energy to an expo-
nential and normalizing:
p(x, h) =
1
Z
exp{−E(x, h)}. (10)
Computing the partition function Z is generally in-
tractable. However, conditional inference in the form of
computing h conditioned on v or vice versa is tractable
and results in a simple formula:
P(h
j
|x) =
1
1 + exp{−b
j
− W
j
x}
. (11)
Since the network is symmetric, a similar expression
holds for P(x
i
|h).
DBNs (Bengio et al., 2007; Hinton et al., 2006) are
essentially SAEs where the AE layers are replaced by
RBMs. Training of the individual layers is, again, done
in an unsupervised manner. Final fine-tuning is per-
formed by adding a linear classifier to the top layer of
the DBN and performing a supervised optimization.
2.6.3. Variational Auto-Encoders and Generative Ad-
verserial Networks
Recently, two novel unsupervised architectures
were introduced: the variational auto-encoder (VAE)
(Kingma and Welling, 2013) and the generative adver-
sarial network (GAN) (Goodfellow et al., 2014). There
are no peer-reviewed papers applying these methods to
medical images yet, but applications in natural images
are promising. We will elaborate on their potential in
the discussion.
2.7. Hardware and Software
One of the main contributors to steep rise of deep
learning has been the widespread availability of GPU
and GPU-computing libraries (CUDA, OpenCL). GPUs
are highly parallel computing engines, which have an
order of magnitude more execution threads than central
processing units (CPUs). With current hardware, deep
learning on GPUs is typically 10 to 30 times faster than
on CPUs.
Next to hardware, the other driving force behind the
popularity of deep learning methods is the wide avail-
ability of open source software packages. These li-
braries provide efficient GPU implementations of im-
portant operations in neural networks, such as convo-
lutions; allowing the user to implement ideas at a high
level rather than worrying about low-level efficient im-
plementations. At the time of writing, the most popular
packages were (in alphabetical order):
• Caffe (Jia et al., 2014). Provides C++ and Python
interfaces, developed by graduate students at UC
Berkeley.
• Tensorflow (Abadi et al., 2016). Provides C++
and Python and interfaces, developed by Google
and is used by Google research.
• Theano (Bastien et al., 2012). Provides a Python
interface, developed by MILA lab in Montreal.
• Torch (Collobert et al., 2011). Provides a Lua in-
terface and is used by, among others, Facebook AI
research.
There are third-party packages written on top of one or
more of these frameworks, such as Lasagne (https://
github.com/Lasagne/Lasagne) or Keras (https:
//keras.io/). It goes beyond the scope of this paper
to discuss all these packages in detail.
3. Deep Learning Uses in Medical Imaging
3.1. Classification
3.1.1. Image/exam classification
Image or exam classification was one of the first ar-
eas in which deep learning made a major contribution
to medical image analysis. In exam classification one
typically has one or multiple images (an exam) as in-
put with a single diagnostic variable as output (e.g.,
disease present or not). In such a setting, every diag-
nostic exam is a sample and dataset sizes are typically
7