6 Lagani, et al.
Note that, more in general, local learning rules with higher order interactions can be considered. A general form for
such a local synaptic update equation, for a generic synaptic connection 𝑖, can be expressed as [61]:
Δ𝑤
𝑖
= 𝑎
0
+ 𝑎
1
𝑥
𝑖
+ 𝑎
2
𝑦 + 𝑎
3
𝑥
𝑖
𝑦 + 𝑎
4
𝑥
2
𝑖
+ 𝑎
5
𝑦
2
+ ... (4)
where the coecients 𝑎
𝑖
may depend on the weights.
When multiple neurons are involved in a complex network, it is desirable that dierent neurons learn to recognize
dierent patterns. In other words, neural activity should pursue a decorrelated form of coding [
51
,
154
], in order to
maximize the amount of information that neurons can transmit. This can be achieved by leveraging the biological
mechanisms of lateral and inhibitory interaction. For example, Eq. 3 can be coupled with competitive learning mechanisms
such as Winner-Takes-All (WTA) [
67
,
178
]: in this case, the neuron with strongest similarity to a given input is chosen
as the winner, and it is the only one allowed to perform a weight update, by virtue of lateral inhibition [
55
]. In this way,
if a similar input will be presented again in the future, the same neuron will be more likely to win again, and dierent
neurons will specialize on dierent clusters. Other forms of lateral interaction can also enable a group of neurons to
extract further principal components form the input (subspace learning) [15, 93, 180].
As a further extension of PCA-based learning methods, Independent Component Analysis (ICA) aims at removing
higher-order correlations from data [
3
,
17
,
74
,
85
,
91
,
162
]. Sparse Coding (SC) [
154
] is another principle, strictly
related to ICA [
153
], from which biologically plausible HaH feature learning networks can be derived, as in the Locally
Competitive Algorithm (LCA) [175].
Hebbian approaches based on competitive [
4
,
99
,
102
,
108
,
137
,
141
,
207
] or subspace learning [
10
,
86
,
104
,
109
] have
recently been subject of signicant interest towards applications in DL contexts.
4 SPIKING NEURAL NETWORKS
This Section discusses models of neural computation based on Spiking Neural Networks (SNNs) [
61
], which more
faithfully resemble real neurons compared to traditional ANN models. We start by introducing the various neuron
models for SNN simulation. We highlight the applications related to biological and neuromorphic computing, which are
of strong practical interest thanks to the energy eciency of the underlying computing paradigm, and we discuss the
challenges related to SNN traning. We describe the biological plasticity models for spiking neurons, and the approaches
to translate backprop-based training to the spiking setting. Finally, an overview of some existing experimental results is
given.
In the following, subsection 4.1 illustrates the main models of spiking neurons and spike-based strategies for
information encoding; subsection 4.2 highlights promising applications of spiking models in the eld of neuromorphic
and biological computing, and the challenges of learning with spiking models; therefore, subsection 4.3 discusses the
plasticity models for spiking neurons, based on Spike Time Dependent Plasticity (STDP); nally, subsection 4.4 presents
some experimental results from literature regarding the applications of SNNs and STDP in DL contexts.
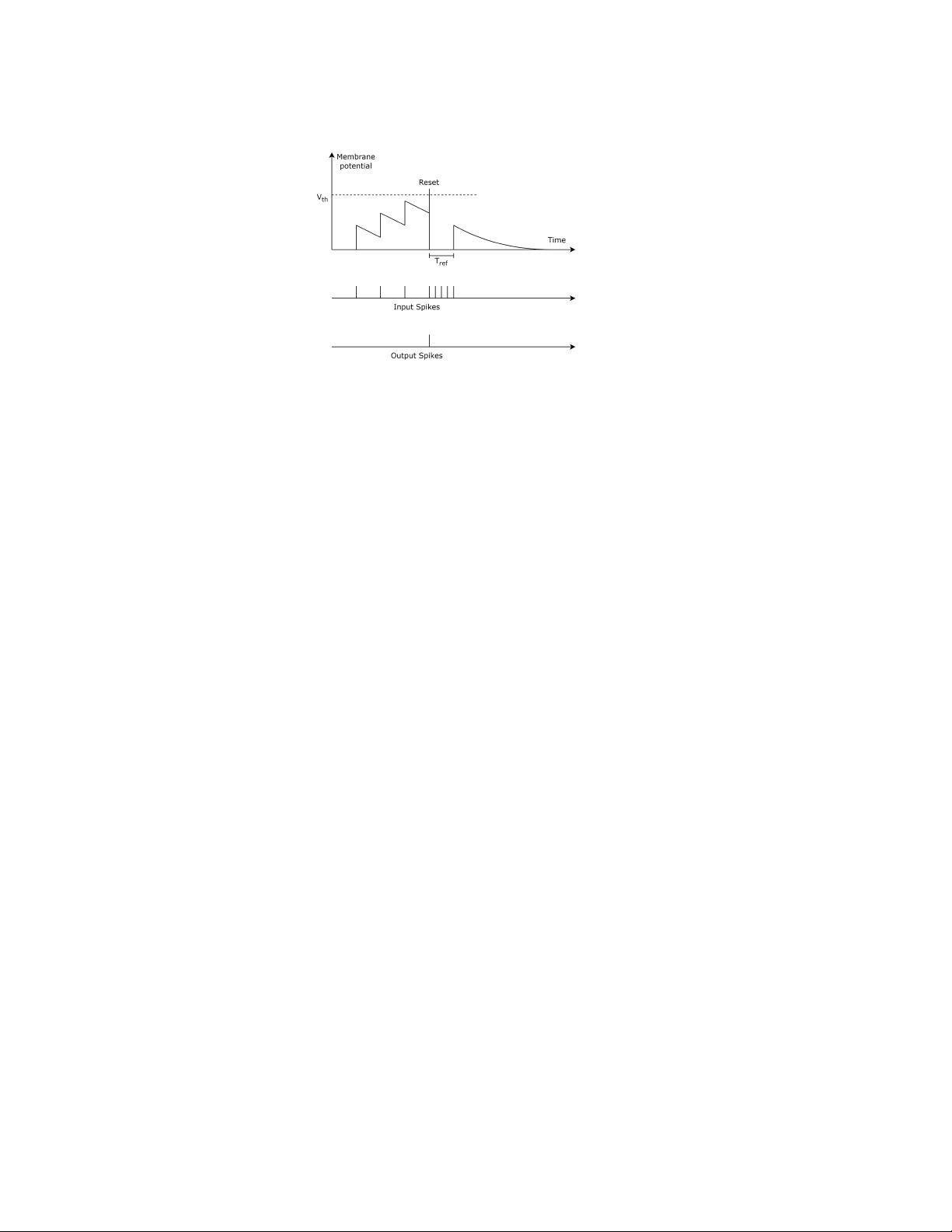
4.1 Spiking Neuron Models and Spike Codes
Spiking Neural Networks (SNNs) are a realistic model of biological networks [
61
,
126
]. While in traditional Articial
Neural Networks (ANNs), neurons communicate via real-valued signals, in SNNs they emit short pulses called spikes.
All the spikes are equal to each other and values are encoded in the timing or in the frequency with which spikes are
emitted.
Various spiking neuron models have been proposed in literature [61], which we highlight in the following:
Manuscript submitted to ACM