samples are covered by more than one module members (called
low impurity). Such properties can be used to infer combin-
ational scores to qualify mutual exclusivity. However, searching
for candidate mutually exclusive modules is a NP-hard problem
[44, 47, 48]. Hundreds, even thousands, of somatic alterations are
detected in certain cancer types, and their possible combinations
become enormous. There are as many as 10
8
–10
11
for combin-
ations of four genes, making exact enumeration impossible.
A feasible resolution to this dilemma is utility of heuristic algo-
rithms, such as the greedy algorithm, the Markov chain Monte
Carlo (MCMC) method and the simulated annealing algorithm
[43, 44].
Vandin et al. [44] developed Dendrix that adopts two search-
ing strategies, including the greedy algorithm and the MCMC
method, to identify mutually exclusive modules. In brief,
starting from seed genes, the greedy algorithm iteratively adds
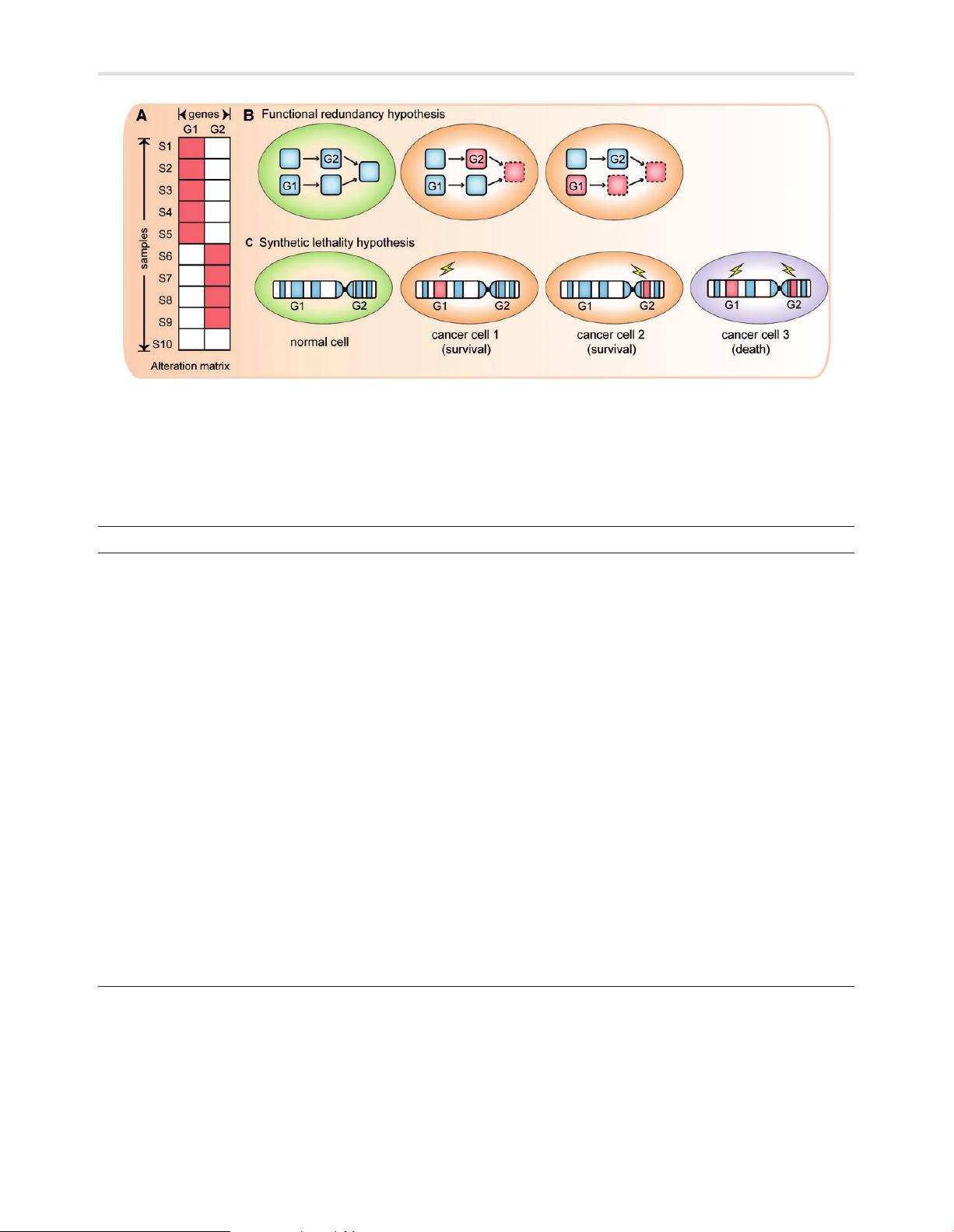
Figure 1. The common hypotheses of mutual exclusivity. (A) Mutual exclusivity between G1 and G2. (B) The functional redundancy hypothesis. Suppose G1 and G2 lie
in the same pathway, and they share the same downstream effect. Squares indicate molecular members in the pathway. While solid squares in red exhibit genetic
alterations, solid squares in blue exhibit normal activity. Dotted squares in red indicate dysfunction molecules in this pathway, which are induced by the genetic
alteration of G1 or G2. (C) The synthetic lethality hypothesis, i.e. the co-occurrence of G1 and G2 is detrimental to cancer cells. Red part shows genetic alteration.
Circles in green indicate normal cells, circles in orange indicate living cancer cells and circles in purple indicate death of cancer cells.
Table 1. Examples of mutual exclusivity with biological significance
Gene 1 Gene 2 Cancer type Phenotype Pubmed ID
BRCA1 CCNE1 Ovary cancer Synthetic lethality 24218601
KRAS TP53 Lung cancer Synthetic lethality 19075675
ATM TP53 Colorectal adenocarcinoma Synthetic lethality 22660439
BRCA2 TP53 HeLa cells; breast cancer Synthetic lethality 17000754
BRCA1 PARP1 Breast cancer Synthetic lethality 21487248
STAG2 STAG1 leukemia, sarcoma, glioblast-
oma and bladder cancer
Synthetic lethality 28430577
BRAF KRAS Colorectal adenocarcinoma;
lung cancer; ovary cancer;
gastric cancer; melanoma;
thyroid cancer; pancreas
cancer; cholangiocarcinoma
MAPK–ERK pathway 21102258; 20645028; 16721043;
19694828; 24959217;
23625203; 26996308;
27823638
PTEN PIK3CA Sarcoma; head and neck can-
cer; breast cancer; brain can-
cer; prostate cancer;
colorectal adenocarcinoma
PI3-kinase/AKT signal-
ing pathway
12569555; 22020193; 18097548;
19443396; 15805248;
ERBB2 KRAS Gastric cancer; lung cancer;
ovary cancer
EGFR/KRAS/BRAF
pathway
22315472; 25047676; 15753357;
17459062; 22899400
CDKN2A TP53 Leukemia; head and neck can-
cer; cholangiocarcinoma
Cell cycle pathway 15896902; 10675493; 18246599;
27495988
ERBB2 EGFR Lung cancer; gastric cancer;
head and neck cancer; brain
cancer
EGFR-RAS-RAF signaling
pathway
25047676; 22042947; 23855785;
17459062; 18757405
BRAF NRAS Thyroid cancer; brain cancer;
melanoma; leukemia; colo-
rectal adenocarcinoma
EGFR-RAS-RAF signaling
pathway
26575603; 21828154; 25576899;
24335104; 18381570;
25848750; 23860532;
16784981
Mutual exclusivity across cancer genomes | 3









