Locally Densest Subgraph Discovery
Lu Qin
†
, Rong-Hua Li
‡
, Lijun Chang
§
, Chengqi Zhang
†
†
Centre for Quantum Computation & Intelligent Systems, University of Technology, Sydney, Australia
‡
Shenzhen University, China
§
The University of New South Wales, Australia
†
{lu.qin,chengqi.zhang}@uts.edu.au
‡
rhli@szu.edu.cn
§
ljchang@cse.unsw.edu.au
ABSTRACT
Mining dense subgraphs from a large graph is a fundamental graph
mining task and can be widely applied in a variety of application
domains such as network science, biology, graph database, web
mining, graph compression, and micro-blogging systems. Here a
dense subgraph is defined as a subgraph with high density (#.edge
/ #.node). Existing studies of this problem either focus on finding
the densest subgraph or identifying an optimal clique-like dense
subgraph, and they adopt a simple greedy approach to find the top-
k dense subgraphs. However, their identified subgraphs cannot be
used to represent the dense regions of the graph. Intuitively, to
represent a dense region, the subgraph identified should be the sub-
graph with highest density in its local region in the graph. However,
it is non-trivial to formally model a locally densest subgraph. In this
paper, we aim to discover top-k such representative locally densest
subgraphs of a graph. We provide an elegant parameter-free defini-
tion of a locally densest subgraph. The definition not only fits well
with the intuition, but is also associated with several nice structural
properties. We show that the set of locally densest subgraphs in
a graph can be computed in polynomial time. We further propose
three novel pruning strategies to largely reduce the search space
of the algorithm. In our experiments, we use several real datasets
with various graph properties to evaluate the effectiveness of our
model using four quality measures and a case study. We also test
our algorithms on several real web-scale graphs, one of which con-
tains 118.14 million nodes and 1.02 billion edges, to demonstrate
the high efficiency of the proposed algorithms.
Categories and Subject Descriptors
G.2.2 [Graph Theory]: Graph Algorithms; H.2.8 [Database Ap-
plications]: Data Mining
Keywords
Graph; Dense Subgraph; Big Data
1. INTRODUCTION
Mining dense subgraphs from a large graph is a fundamental
graph mining task which has been widely used in a variety of
Singhal
John M.
Prager
Wessel
Kraaij
Christ
Buckley
Alan F.
Smeaton
David J.
Harper
Thomas
Hofmann
Susan T.
Dumais
Mark
Sanderson
Norbert
Fuhr
Donna
Harman
David D.
Lewis
Jamie
Callan
Eric
Horvitz
Simon
Parsons
Rudolf
Kruse
Philippe
Smets
Serafin
Moral
Didier
Dubois
Henri
Prade
W. Bruce
Croft
Hector
Geffner
Gregory
Cooper
IR
*
BN
G
BN
G
IR
G
G
IR
Amit
G’
G
Jrme Lang
Jinxi Xu
Allan
*
James
v
24
v
23
v
v
21
v
20
v
19
v
18
v
17
v
16
v
15
v
14
v
13
v
12
v
10
v
9
v
8
v
7
v
6
v
5
v
4
v
3
v
25
v
2
v
26
v
1
v
11
22
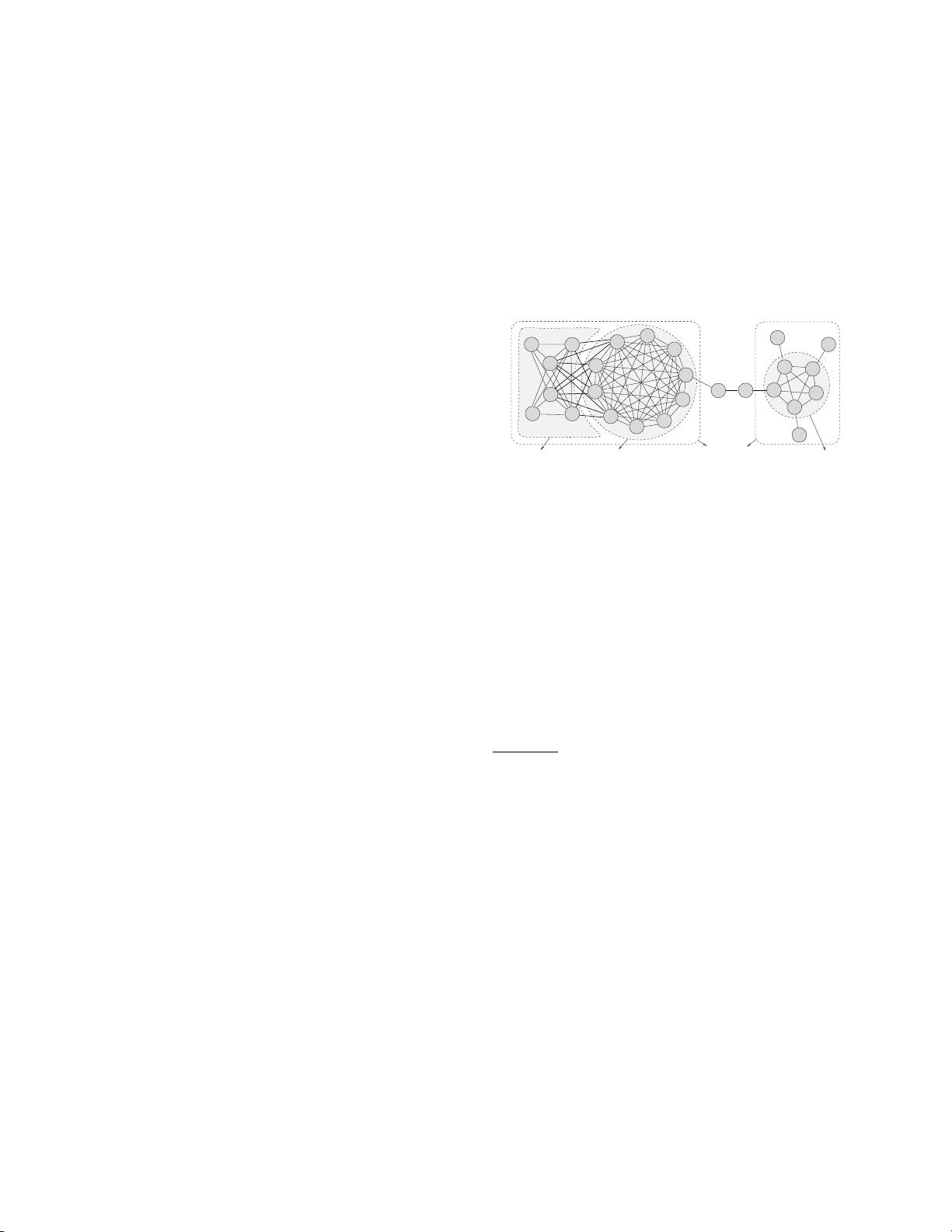
Figure 1: Part of the Coauthor Network
application domains [28]. For example, in the network science do-
main, dense subgraphs represent cohesive groups or communities
in a network. There are several community detection algorithms
that are based on dense subgraphs [20, 14]. In the biology domain,
the dense subgraph mining problem has been leveraged to iden-
tify regulatory motifs in genomic DNA [21] and to find complex
patterns in a gene annotation graph [31]. In the graph database
domain, algorithms for dense subgraphs discovery play an impor-
tant role for creating elegant index structures to process reachability
and distance queries efficiently [17, 26]. In the web mining do-
main, dense subgraph mining techniques are applied to link spam
detection based on an interesting observation that dense subgraphs
typically correspond to link spam farms [23]. In addition, dense
subgraph mining has also been used for graph compression [11]
and identifying stories in micro-blogging systems [4].
Motivation
. The dense subgraph mining problem aims at identify-
ing the subgraphs with high density (i.e., #.edge / #.node) [25, 6,
13, 7] from a large graph. Existing studies of this problem either
focus on finding the densest subgraph (the subgraph with the high-
est density) [25, 6] or identifying an optimal clique-like dense sub-
graph (e.g., optimal quasi-clique proposed in [37]). To find top-k
dense subgraphs, a simple greedy procedure, as suggested in [37],
is used, which iteratively invokes the same algorithm k times in
the residual graph after deleting the identified dense subgraphs in
the previous iterations. The major drawback of these methods is
that their results cannot be used to represent the dense regions of
the graph. If the graph contains a large dense region, the top-k
dense subgraphs identified by the above approaches may all be-
long to the same dense region, and other local dense regions may
be neglected. For instance, Fig. 1 shows part of the collaboration
network in the Coauthor dataset (http://arnetminer.org/), which in-
cludes two subgraphs G
IR
and G
BN
in two research areas Informa-
tion Retrieval (IR) and Bayesian Networks (BN) respectively. If we
use the greedy procedure to find the top-2 dense subgraphs based
on either the densest subgraph model [25] or the optimal quasi-
clique model [37], the result will be G
∗
IR
and G
0
IR
. Intuitively, the
two dense subgraphs cannot fully reflect the top-2 representative
dense regions of the graph, because G
0
IR
is located in the same
Permission to make digital or hard copies of all or part of this work for personal
or classroom use is granted without fee provided that copies are not made or
distributed for profit or commercial advantage and that copies bear this notice
and the full citation on the first page. Copyrights for components of this work
owned by others than ACM must be honored. Abstracting with credit is
permitted. To copy otherwise, or republish, to post on servers or to redistribute
to lists, requires prior specific permission and/or a fee. Request permissions
from Permissions@acm.org.
KDD '15, August 10-13, 2015, Sydney, NSW, Australia.
© 2015 ACM. ISBN 978-1-4503-3664-2/15/08…$15.00.
DOI: http://dx.doi.org/10.1145/2783258.2783299









