A LiveCoMS Tutorial
emstep = 0.01
nsteps = 50000
These instructions tell the GROMACS program
mdrun
(dis-
cussed below) to use the steepest descents method for en-
ergy minimization, and to terminate the process if the magni-
tude of the potential energy gradient is 1000.0 kJ mol
-1
nm
-1
or smaller. The maximum step size along the gradient is 0.01
nm, and a maximum of 50000 steps are allowed. The default
value of
emstep
in GROMACS of 0.01 nm is used, but it is often
necessary to use a smaller maximum step (e.g. 0.002 nm)
for systems that have difficulty converging; the smaller step
size allows for a more thorough walk over the potential en-
ergy surface, whereas a larger step may miss a path to the
true minimum. It is also possible to allow the process to go
on indefinitely, stopping only when convergence is reached
(due to numerical precision or by actually achieving
emtol
) by
setting
nsteps = -1
. Note that
emstep
has units of distance,
not time. Energy minimization is not a dynamical process;
time does not elapse between each step and there are no
velocities. Energy minimization steps are expressed in terms
of maximum displacement along the vector indicated by the
force.
The remainder of the
.mdp
file contains instructions for
how to compute nonbonded interactions:
ns tlist = 1
cutoff - sche me = Verlet
ns _type = grid
cou l o m b type = PME
rc oulomb = 1.0
rvdw = 1.0
pbc = xyz
The value of
nstlist
controls how many steps (minimization
or MD integration) elapse between updating the neighbor list
for determining which atoms contribute to the short-range
forces. In MD simulations, this value is larger because atoms
exchange from the neighbor list in diffusion-limited time, but
for energy minimization, it needs to be set to 1 because
the configuration may change considerably between each
step. The
cutoff-scheme = Verlet
setting uses a buffered
neighbor list, that is, atoms outside the longest cutoff are still
tracked to improve energy conservation. Neighbor searching
is done by checking atoms in neighboring grids (
ns_type =
grid
) rather than checking every possible atom (
ns_type =
simple
). All short-range nonbonded interactions (electrostat-
ics and van der Waals) are truncated at 1.0 nm (
rcoulomb =
1.0
and
rvdw= 1.0
), and periodic boundary conditions are
applied in all three dimensions (
pbc = xyz
). The PME method
is used to calculate long-range electrostatic forces.
Note that in
.mdp
files, there is no difference between
a hyphen (-) and underscore (_); hence in the above exam-
ple,
ns-type
and
ns_type
would be equivalent, as would
cutoff-scheme and cutoff_scheme.
As above, invoke
grompp
to assemble the instructions for
energy minimization (-f minim.mdp), coordinates
(
-c 1AKI_solv_ions.gro
), and topology (
-p topol.top
) to
create the run input file for energy minimization (
-o em.tpr
):
$ gmx gromp p -f minim . mdp -c
1 A K I _ s o l v _ i o n s . gro -p topol . top -o
em . tpr
The GROMACS
mdrun
program is responsible for perform-
ing all minimization and dynamics processes. To run energy
minimization, use the following syntax:
$ gmx mdrun -v - deffnm em
The
-v
option invokes "verbose" mode, in which an estimate
of time remaining is printed to the screen, along with the
current energy minimization step, the step size, potential
energy, and maximum force. It is not a required option, and if
printed to a file, may result in a large file that is saved to disk.
It is, however, instructive to watch the progress of energy
minimization to understand what is going on. The
-deffnm
option defines the base file name for all input and output
files and eliminates the need for using individual input and
output flags to specify names.
mdrun
requires the
.tpr
file as its only input, normally
passed to the
-s
flag.
mdrun
produces several file types, in-
cluding an ASCII text file with a
.log
extension, a binary file
with all energy values with a
.edr
extension, and trajectory
files with either
.trr
(full-precision coordinates, velocities,
and/or forces) or
.xtc
(reduced precision coordinates only)
extensions. When using
-deffnm
, these files will be called
em
,
with a corresponding file extension. It is useful to name files
in this manner to avoid relying on default file names (
md.log
,
ener.edr
,
traj.trr
,
traj_comp.xtc
), which will be the same
for any process carried out by mdrun.
When
mdrun
is done, the user will see something similar
to the following:
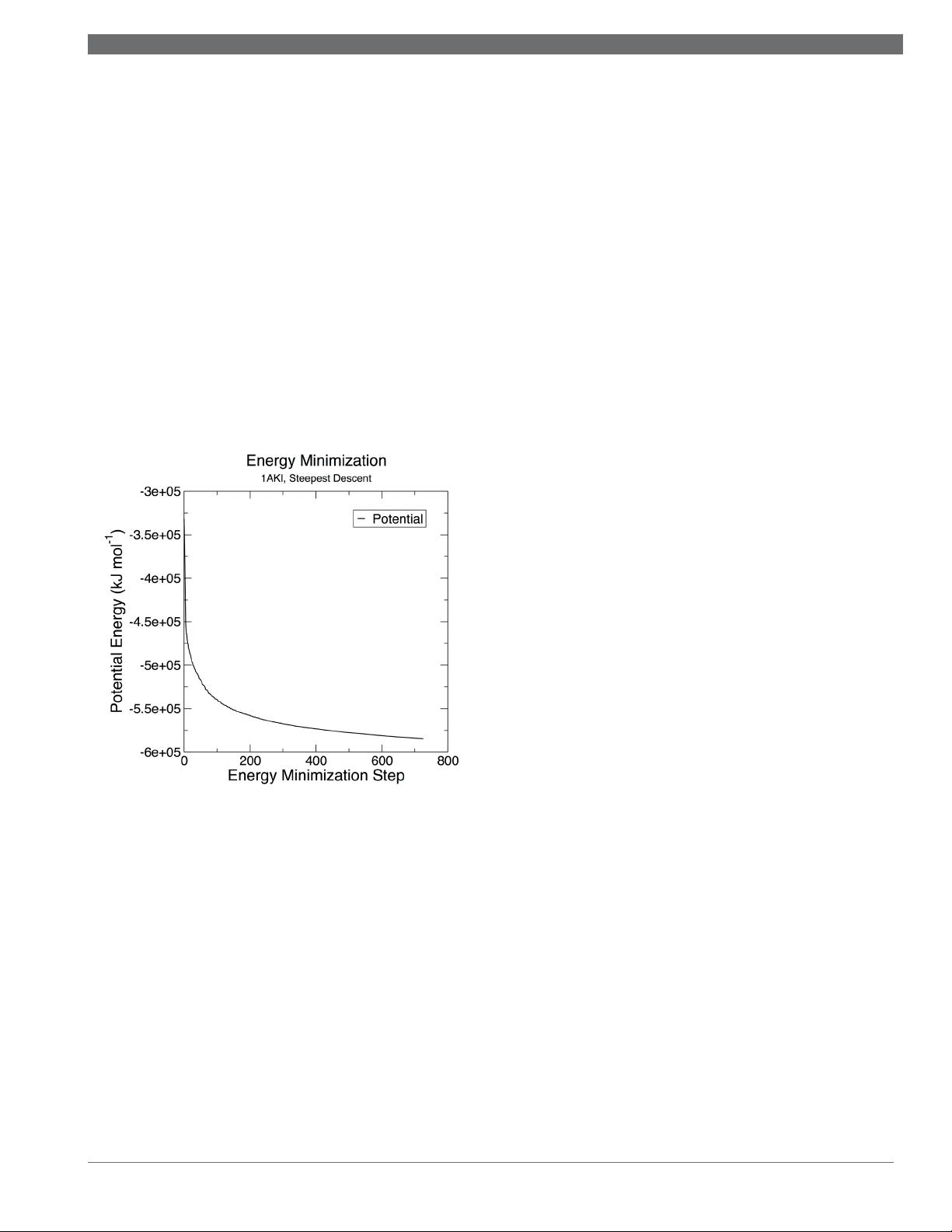
Stee p est D e scen t s co n verg e d to Fmax < 1000 i n 726 step s
Pote n tia l Ene r g y = -5. 8 4 4 8 7 69 e +05
Maxim u m force = 9.69 5 7593 e +02 on atom 736
Norm of f o r c e = 2 . 3290 9 28 e +01
For a system such as this, it is expected that the final po-
tential energy will be a negative value, on the order of 10
5
- 10
6
kJ mol
-1
. Potential energy is an extrinsic quantity, so
larger systems will have larger magnitudes. For a solvated
protein, the value will be negative, as favorable electrostatic
interactions between water molecules dominate the poten-
tial energy terms. Smaller systems may even have positive
potential energy, arising from the fact that in such cases, the
intramolecular bonded terms have a larger magnitude than
9 of 53
https://doi.org/10.33011/livecoms.1.1.5068
Living J. Comp. Mol. Sci. 2019, 1(1), 5068